Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal
- PMID: 21685894
- PMCID: PMC3129424
- DOI: 10.1038/ncb2283
Opposing effects of Tcf3 and Tcf1 control Wnt stimulation of embryonic stem cell self-renewal
Abstract
The co-occupancy of Tcf3 with Oct4, Sox2 and Nanog on embryonic stem cell (ESC) chromatin indicated that Tcf3 has been suggested to play an integral role in a poorly understood mechanism underlying Wnt-dependent stimulation of mouse ESC self-renewal of mouse ESCs. Although the conventional view of Tcf proteins as the β-catenin-binding effectors of Wnt signalling suggested Tcf3-β-catenin activation of target genes would stimulate self-renewal, here we show that an antagonistic relationship between Wnt3a and Tcf3 on gene expression regulates ESC self-renewal. Genetic ablation of Tcf3 replaced the requirement for exogenous Wnt3a or GSK3 inhibition for ESC self-renewal, demonstrating that inhibition of Tcf3 repressor is the necessary downstream effect of Wnt signalling. Interestingly, both Tcf3-β-catenin and Tcf1-β-catenin interactions contributed to Wnt stimulation of self-renewal and gene expression, and the combination of Tcf3 and Tcf1 recruited Wnt-stabilized β-catenin to Oct4 binding sites on ESC chromatin. This work elucidates the molecular link between the effects of Wnt and the regulation of the Oct4/Sox2/Nanog network.
Figures
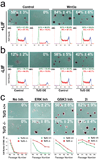
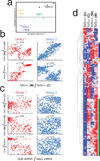

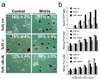
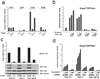
Comment in
-
Wnt: what's needed to maintain pluripotency?Nat Cell Biol. 2011 Sep 2;13(9):1024-6. doi: 10.1038/ncb2333. Nat Cell Biol. 2011. PMID: 21892143
References
-
- Liang J, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10:731–739. - PubMed
-
- Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006 - PubMed
-
- Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

