A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease
- PMID: 21685905
- PMCID: PMC3366591
- DOI: 10.1038/nbt.1899
A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease
Abstract
We developed a pipeline to integrate the proteomic technologies used from the discovery to the verification stages of plasma biomarker identification and applied it to identify early biomarkers of cardiac injury from the blood of patients undergoing a therapeutic, planned myocardial infarction (PMI) for treatment of hypertrophic cardiomyopathy. Sampling of blood directly from patient hearts before, during and after controlled myocardial injury ensured enrichment for candidate biomarkers and allowed patients to serve as their own biological controls. LC-MS/MS analyses detected 121 highly differentially expressed proteins, including previously credentialed markers of cardiovascular disease and >100 novel candidate biomarkers for myocardial infarction (MI). Accurate inclusion mass screening (AIMS) qualified a subset of the candidates based on highly specific, targeted detection in peripheral plasma, including some markers unlikely to have been identified without this step. Analyses of peripheral plasma from controls and patients with PMI or spontaneous MI by quantitative multiple reaction monitoring mass spectrometry or immunoassays suggest that the candidate biomarkers may be specific to MI. This study demonstrates that modern proteomic technologies, when coherently integrated, can yield novel cardiovascular biomarkers meriting further evaluation in large, heterogeneous cohorts.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Figures



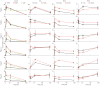
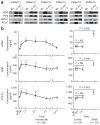
Comment in
-
Streamlining biomarker discovery.Nat Biotechnol. 2011 Jul 11;29(7):600-2. doi: 10.1038/nbt.1917. Nat Biotechnol. 2011. PMID: 21747385 No abstract available.
References
-
- Edwards AVG, White MY, Cordwell SJ. The role of proteomics in clinical cardiovascular biomarker discovery. Mol Cell Proteomics. 2008;7:1824–1837. - PubMed
-
- Fu Q, Van Eyk JE. Proteomics and heart disease: identifying biomarkers of clinical utility. Expert Rev Proteomics. 2006;3:237–249. - PubMed
-
- Anderson NL. The clinical plasma proteome: a survey of clinical assays for proteins in plasma and serum. Clin Chem. 2010;56:177–185. - PubMed
-
- Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

