CTCF-mediated functional chromatin interactome in pluripotent cells
- PMID: 21685913
- PMCID: PMC3436933
- DOI: 10.1038/ng.857
CTCF-mediated functional chromatin interactome in pluripotent cells
Erratum in
- Nat Genet. 2011 Aug;43(8):815
Abstract
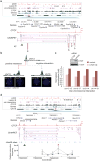
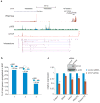
Mammalian genomes are viewed as functional organizations that orchestrate spatial and temporal gene regulation. CTCF, the most characterized insulator-binding protein, has been implicated as a key genome organizer. However, little is known about CTCF-associated higher-order chromatin structures at a global scale. Here we applied chromatin interaction analysis by paired-end tag (ChIA-PET) sequencing to elucidate the CTCF-chromatin interactome in pluripotent cells. From this analysis, we identified 1,480 cis- and 336 trans-interacting loci with high reproducibility and precision. Associating these chromatin interaction loci with their underlying epigenetic states, promoter activities, enhancer binding and nuclear lamina occupancy, we uncovered five distinct chromatin domains that suggest potential new models of CTCF function in chromatin organization and transcriptional control. Specifically, CTCF interactions demarcate chromatin-nuclear membrane attachments and influence proper gene expression through extensive cross-talk between promoters and regulatory elements. This highly complex nuclear organization offers insights toward the unifying principles that govern genome plasticity and function.
Figures




Comment in
-
Mapping higher order structure of chromatin domains.Nat Genet. 2011 Jun 28;43(7):615-6. doi: 10.1038/ng.869. Nat Genet. 2011. PMID: 21709679 No abstract available.
References
-
- Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–43. - PubMed
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
-
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540–6. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

