Identification of a novel PEX14 mutation in Zellweger syndrome
- PMID: 21686775
- PMCID: PMC3027610
- DOI: 10.1136/bcr.07.2008.0503
Identification of a novel PEX14 mutation in Zellweger syndrome
Abstract
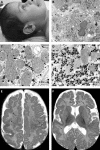
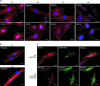
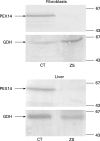
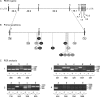
Here we report a patient with Zellweger syndrome, who presented at the age of 3 months with icterus, dystrophy, axial hypotonia, and hepatomegaly. Abnormal findings of metabolic screening tests included hyperbilirubinaemia, hypoketotic dicarboxylic aciduria, increased C(26:0) and decreased C(22:0) plasma levels, and strongly reduced plasmalogen concentrations. In fibroblasts, both peroxisomal α- and β-oxidation were impaired. Liver histology revealed bile duct paucity, cholestasis, arterial hyperplasia, very small branches of the vena portae, and parenchymatic destruction. Immunocytochemical analysis of cultured fibroblasts demonstrated that the cells contain peroxisomal remnants lacking apparent matrix protein content and PEX14, a central membrane component of the peroxisomal matrix protein import machinery. Transfection of fibroblasts with a plasmid coding for wild-type PEX14 restored peroxisomal matrix protein import. Mutational analysis of this gene revealed a genomic deletion leading to the deletion of exon 3 from the coding DNA (c.85-?_170+?del) and a concomitant change of the reading frame (p.[Ile29_Lys56del;Gly57GlyfsX2]).
Figures
References
-
- Mannaerts GP, Van Veldhoven PP, Casteels M. Peroxisomal lipid degradation via beta- and alpha-oxidation in mammals. Cell Biochem Biophys 2003; 32: 73–87 - PubMed
-
- Wanders RJ, Waterham HR. Peroxisomal disorders: the single peroxisomal enzyme deficiencies. Biochim Biophys Acta 2006; 1763: 1707–20 - PubMed
-
- Shimozawa N. Molecular and clinical aspects of peroxisomal diseases. J Inherit Metab Dis 2007; 30: 193–7 - PubMed
-
- Steinberg SJ, Dodt G, Raymond GV, et al. Peroxisome biogenesis disorders. Biochim Biophys Acta 2006; 1763: 1733–48 - PubMed
-
- Goldfischer S, Moore CL, Johnson AB, et al. Peroxisomal and mitochondrial defects in the cerebro-hepato-renal syndrome. Science 1973; 182: 62–4 - PubMed
LinkOut - more resources
Full Text Sources
