Genetic and Functional Diversity of Pseudomonas aeruginosa Lipopolysaccharide
- PMID: 21687428
- PMCID: PMC3108286
- DOI: 10.3389/fmicb.2011.00118
Genetic and Functional Diversity of Pseudomonas aeruginosa Lipopolysaccharide
Abstract
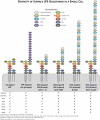
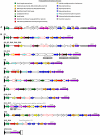
Lipopolysccharide (LPS) is an integral component of the Pseudomonas aeruginosa cell envelope, occupying the outer leaflet of the outer membrane in this Gram-negative opportunistic pathogen. It is important for bacterium-host interactions and has been shown to be a major virulence factor for this organism. Structurally, P. aeruginosa LPS is composed of three domains, namely, lipid A, core oligosaccharide, and the distal O antigen (O-Ag). Most P. aeruginosa strains produce two distinct forms of O-Ag, one a homopolymer of D-rhamnose that is a common polysaccharide antigen (CPA, formerly termed A band), and the other a heteropolymer of three to five distinct (and often unique dideoxy) sugars in its repeat units, known as O-specific antigen (OSA, formerly termed B band). Compositional differences in the O units among the OSA from different strains form the basis of the International Antigenic Typing Scheme for classification via serotyping of different strains of P. aeruginosa. The focus of this review is to provide state-of-the-art knowledge on the genetic and resultant functional diversity of LPS produced by P. aeruginosa. The underlying factors contributing to this diversity will be thoroughly discussed and presented in the context of its contributions to host-pathogen interactions and the control/prevention of infection.
Keywords: bacteriophage; biosynthesis; lipopolysaccharide; motility; nucleotide sugars; seroconversion; serotyping; virulence.
Figures




References
-
- Abeyrathne P. D., Daniels C., Poon K. K., Matewish M. J., Lam J. S. (2005). Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187, 3002–3012 10.1128/JB.187.9.3002-3012.2005 - DOI - PMC - PubMed
-
- Abeyrathne P. D., Lam J. S. (2007). WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core. Mol. Microbiol. 65, 1345–1359 - PubMed
-
- Anderson P. (2010). Emerging therapies in cystic fibrosis. Ther. Adv. Respir. Dis. 4, 177–185 - PubMed
-
- Arsenault T. L., Hughes D. W., Maclean D. B., Szarek W. A., Kropinski A. M. B., Lam J. S. (1991). Structural studies on the polysaccharide portion of ‘A-band’ lipopolysaccharide from a mutant (AK14O1) of Pseudomonas aeruginosa PAO1. Can. J. Chem. 69, 1273–1280
-
- Augustin D. K., Song Y., Baek M. S., Sawa Y., Singh G., Taylor B., Rubio-Mills A., Flanagan J. L., Wiener-Kronish J. P., Lynch S. V. (2007). Presence or absence of lipopolysaccharide O antigens affects type III secretion by Pseudomonas aeruginosa. J. Bacteriol. 189, 2203–2209 10.1128/JB.01839-06 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

