Polymorphic structures of Alzheimer's β-amyloid globulomers
- PMID: 21687730
- PMCID: PMC3110195
- DOI: 10.1371/journal.pone.0020575
Polymorphic structures of Alzheimer's β-amyloid globulomers
Abstract
Background: Misfolding and self-assembly of Amyloid-β (Aβ) peptides into amyloid fibrils is pathologically linked to the development of Alzheimer's disease. Polymorphic Aβ structures derived from monomers to intermediate oligomers, protofilaments, and mature fibrils have been often observed in solution. Some aggregates are on-pathway species to amyloid fibrils, while the others are off-pathway species that do not evolve into amyloid fibrils. Both on-pathway and off-pathway species could be biologically relevant species. But, the lack of atomic-level structural information for these Aβ species leads to the difficulty in the understanding of their biological roles in amyloid toxicity and amyloid formation.
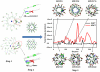
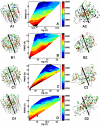
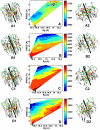
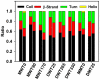
Methods and findings: Here, we model a series of molecular structures of Aβ globulomers assembled by monomer and dimer building blocks using our peptide-packing program and explicit-solvent molecular dynamics (MD) simulations. Structural and energetic analysis shows that although Aβ globulomers could adopt different energetically favorable but structurally heterogeneous conformations in a rugged energy landscape, they are still preferentially organized by dynamic dimeric subunits with a hydrophobic core formed by the C-terminal residues independence of initial peptide packing and organization. Such structural organizations offer high structural stability by maximizing peptide-peptide association and optimizing peptide-water solvation. Moreover, curved surface, compact size, and less populated β-structure in Aβ globulomers make them difficult to convert into other high-order Aβ aggregates and fibrils with dominant β-structure, suggesting that they are likely to be off-pathway species to amyloid fibrils. These Aβ globulomers are compatible with experimental data in overall size, subunit organization, and molecular weight from AFM images and H/D amide exchange NMR.
Conclusions: Our computationally modeled Aβ globulomers provide useful insights into structure, dynamics, and polymorphic nature of Aβ globulomers which are completely different from Aβ fibrils, suggesting that these globulomers are likely off-pathway species and explaining the independence of the aggregation kinetics between Aβ globulomers and fibrils.
Conflict of interest statement
Figures



Similar articles
-
Structural, morphological, and kinetic studies of β-amyloid peptide aggregation on self-assembled monolayers.Phys Chem Chem Phys. 2011 Sep 7;13(33):15200-10. doi: 10.1039/c1cp21156k. Epub 2011 Jul 19. Phys Chem Chem Phys. 2011. PMID: 21769359
-
Role of water in protein aggregation and amyloid polymorphism.Acc Chem Res. 2012 Jan 17;45(1):83-92. doi: 10.1021/ar2000869. Epub 2011 Jul 15. Acc Chem Res. 2012. PMID: 21761818 Free PMC article.
-
The Alzheimer's amyloid-β(1-42) peptide forms off-pathway oligomers and fibrils that are distinguished structurally by intermolecular organization.J Mol Biol. 2013 Jul 24;425(14):2494-508. doi: 10.1016/j.jmb.2013.04.003. Epub 2013 Apr 11. J Mol Biol. 2013. PMID: 23583777 Free PMC article.
-
Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review.
-
Distinct role of hydration water in protein misfolding and aggregation revealed by fluctuating thermodynamics analysis.Acc Chem Res. 2015 Apr 21;48(4):956-65. doi: 10.1021/acs.accounts.5b00032. Epub 2015 Apr 6. Acc Chem Res. 2015. PMID: 25844814 Review.
Cited by
-
Coupling of Zinc-Binding and Secondary Structure in Nonfibrillar Aβ40 Peptide Oligomerization.J Chem Inf Model. 2015 Jun 22;55(6):1218-30. doi: 10.1021/acs.jcim.5b00063. Epub 2015 Jun 3. J Chem Inf Model. 2015. PMID: 26017140 Free PMC article.
-
A randomized, double-blind, phase 2b proof-of-concept clinical trial in early Alzheimer's disease with lecanemab, an anti-Aβ protofibril antibody.Alzheimers Res Ther. 2021 Apr 17;13(1):80. doi: 10.1186/s13195-021-00813-8. Alzheimers Res Ther. 2021. PMID: 33865446 Free PMC article. Clinical Trial.
-
Toxic species in amyloid disorders: Oligomers or mature fibrils.Ann Indian Acad Neurol. 2015 Apr-Jun;18(2):138-45. doi: 10.4103/0972-2327.144284. Ann Indian Acad Neurol. 2015. PMID: 26019408 Free PMC article. Review.
-
Molecular interactions of Alzheimer amyloid-β oligomers with neutral and negatively charged lipid bilayers.Phys Chem Chem Phys. 2013 Jun 21;15(23):8878-89. doi: 10.1039/c3cp44448a. Epub 2013 Mar 14. Phys Chem Chem Phys. 2013. PMID: 23493873 Free PMC article.
-
Structures and dynamics of β-barrel oligomer intermediates of amyloid-beta16-22 aggregation.Biochim Biophys Acta Biomembr. 2018 Sep;1860(9):1687-1697. doi: 10.1016/j.bbamem.2018.03.011. Epub 2018 Mar 14. Biochim Biophys Acta Biomembr. 2018. PMID: 29550287 Free PMC article.
References
-
- Stehanie R, Olivier S, Olivier S, Annick T, Robert B, et al. Fusogenic Alzheimer's peptide fragment Abeta (29–42) in interaction with lipid bilayers: Secondary structure, dynamics, and specific interaction with phosphatidyl ethanolamine polar heads as revealed by solid-state NMR. Protein Sci. 2005;14:1181–1189. - PMC - PubMed
-
- Di Carlo M. Beta amyloid peptide: from different aggregation forms to the activation of different biochemical pathways. Euro Biophys J 2009 - PubMed
-
- Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: The emerging role of oligomeric assemblies. J Neuroscience Research. 2002;69:567–577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

