Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei
- PMID: 21689430
- PMCID: PMC3135491
- DOI: 10.1186/1749-8104-6-29
Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei
Abstract
Background: The ventral midbrain contains a diverse array of neurons, including dopaminergic neurons of the ventral tegmental area (VTA) and substantia nigra (SN) and neurons of the red nucleus (RN). Dopaminergic and RN neurons have been shown to arise from ventral mesencephalic precursors that express Sonic Hedgehog (Shh). However, Shh expression, which is initially confined to the mesencephalic ventral midline, expands laterally and is then downregulated in the ventral midline. In contrast, expression of the Hedgehog target gene Gli1 initiates in the ventral midline prior to Shh expression, but after the onset of Shh expression it is expressed in precursors lateral to Shh-positive cells. Given these dynamic gene expression patterns, Shh and Gli1 expression could delineate different progenitor populations at distinct embryonic time points.
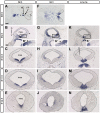
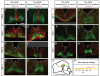
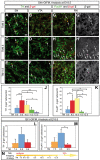
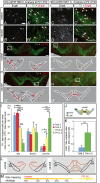
Results: We employed genetic inducible fate mapping (GIFM) to investigate whether precursors that express Shh (Shh-GIFM) or transduce Shh signaling (Gli1-GIFM) at different time points give rise to different ventral midbrain cell types. We find that precursors restricted to the ventral midline are labeled at embryonic day (E)7.5 with Gli1-GIFM, and with Shh-GIFM at E8.5. These precursors give rise to all subtypes of midbrain dopaminergic neurons and the anterior RN. A broader domain of progenitors that includes the ventral midline is marked with Gli1-GIFM at E8.5 and with Shh-GIFM at E9.5; these fate-mapped cells also contribute to all midbrain dopaminergic subtypes and to the entire RN. In contrast, a lateral progenitor domain that is labeled with Gli1-GIFM at E9.5 and with Shh-GIFM at E11.5 has a markedly reduced potential to give rise to the RN and to SN dopaminergic neurons, and preferentially gives rise to the ventral-medial VTA. In addition, cells derived from Shh- and Gli1-expressing progenitors located outside of the ventral midline give rise to astrocytes.
Conclusions: We define a ventral midbrain precursor map based on the timing of Gli1 and Shh expression, and suggest that the diversity of midbrain dopaminergic neurons is at least partially determined during their precursor stage when their medial-lateral position, differential gene expression and the time when they leave the ventricular zone influence their fate decisions.
Figures



References
-
- Ono Y, Nakatani T, Sakamoto Y, Mizuhara E, Minaki Y, Kumai M, Hamaguchi A, Nishimura M, Inoue Y, Hayashi H, Takahashi J, Imai T. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

