Long dwell-time passage of DNA through nanometer-scale pores: kinetics and sequence dependence of motion
- PMID: 21689531
- PMCID: PMC3123917
- DOI: 10.1016/j.bpj.2011.05.007
Long dwell-time passage of DNA through nanometer-scale pores: kinetics and sequence dependence of motion
Abstract
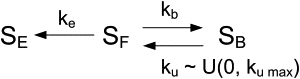
A detailed understanding of the kinetics of DNA motion though nanometer-scale pores is important for the successful development of many of the proposed next-generation rapid DNA sequencing and analysis methods. Many of these approaches require DNA motion through nanopores to be slowed by several orders of magnitude from its native translocation velocity so that the translocation times for individual nucleotides fall within practical timescales for detection. With the increased dwell time of DNA in the pore, DNA-pore interactions begin to play an increasingly important role in translocation kinetics. In previous work, we and others observed that when the DNA dwell time in the pore is substantial (>1 ms), DNA motion in α-hemolysin (α-HL) pores leads to nonexponential kinetics in the escape of DNA out of the pore. Here we show that a three-state model for DNA escape, involving stochastic binding interactions of DNA with the pore, accurately reproduces the experimental data. In addition, we investigate the sequence dependence of the DNA escape process and show that the interaction strength of adenine with α-HL is substantially lower relative to cytosine. Our results indicate a difference in the process by which DNA moves through an α-HL nanopore when the motion is fast (microsecond timescale) as compared with when it is slow (millisecond timescale) and strongly influenced by DNA-pore interactions of the kind reported here. We also show the ability of wild-type α-HL to detect and distinguish between 5-methylcytosine and cytosine based on differences in the absolute ionic current through the pore in the presence of these two nucleotides. The results we present here regarding sequence-dependent (and dwell-time-dependent) DNA-pore interaction kinetics will have important implications for the design of methods for DNA analysis through reduced-velocity motion in nanopores.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bayley H. Sequencing single molecules of DNA. Curr. Opin. Chem. Biol. 2006;10:628–637. - PubMed
-
- de Zoysa R.S.S., Jayawardhana D.A., Guan X. Slowing DNA translocation through nanopores using a solution containing organic salts. J. Phys. Chem. B. 2009;113:13332–13336. - PubMed
-
- Tabard-Cossa V., Trivedi D., Marziali A. Noise analysis and reduction in solid-state nanopores. Nanotechnology. 2007;18:305505.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

