Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas
- PMID: 21690253
- PMCID: PMC3135371
- DOI: 10.1084/jem.20110283
Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas
Abstract
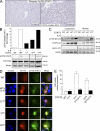
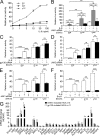
Inflammatory hepatocellular adenomas (IHCAs) are benign liver tumors. 60% of these tumors have IL-6 signal transducer (IL6ST; gp130) mutations that activate interleukin 6 (IL-6) signaling. Here, we report that 12% of IHCA subsets lacking IL6ST mutations harbor somatic signal transducer and activator of transcription 3 (STAT3) mutations (6/49). Most of these mutations are amino acid substitutions in the SH2 domain that directs STAT3 dimerization. In contrast to wild-type STAT3, IHCA STAT3 mutants constitutively activated the IL-6 signaling pathway independent of ligand in hepatocellular cells. Indeed, the IHCA STAT3 Y640 mutant homodimerized independent of IL-6 and was hypersensitive to IL-6 stimulation. This was associated with phosphorylation of tyrosine 705, a residue required for IL-6-induced STAT3 activation. Silencing or inhibiting the tyrosine kinases JAK1 or Src, which phosphorylate STAT3, impaired constitutive activity of IHCA STAT3 mutants in hepatocellular cells. Thus, we identified for the first time somatic STAT3 mutations in human tumors, revealing a new mechanism of recurrent STAT3 activation and underscoring the role of the IL-6-STAT3 pathway in benign hepatocellular tumorigenesis.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

