Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection
- PMID: 21690298
- PMCID: PMC3147801
- DOI: 10.1128/MCB.05073-11
Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection
Abstract
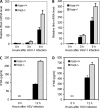
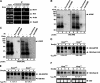
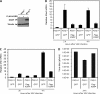
Inherently unstable mRNAs contain AU-rich elements (AREs) in the 3' untranslated regions. Expression of ARE-containing type I interferon transcripts is robustly induced upon viral infection and rapidly shut off thereafter. Their transient accumulation is partly mediated through posttranscriptional regulation. Here we show that mouse embryonic fibroblasts derived from knockout mice deficient in KH-type splicing regulatory protein (KSRP), an RNA-binding protein required for ARE-mediated mRNA decay, produce higher levels of Ifna and Ifnb mRNAs in response to viral infection as a result of decreased mRNA decay. Functional analysis showed that KSRP is required for the decay of Ifna4 and Ifnb mRNAs by interaction with AREs. The increased IFN expression renders Ksrp(-)(/)(-) cells refractory to herpes simplex virus type 1 and vesicular stomatitis virus infection. These findings support a role of a posttranscriptional mechanism in the control of type I IFN expression and highlight the function of KSRP in innate immunity by negatively regulating IFN production.
Figures


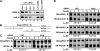


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
