Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods
- PMID: 21693555
- PMCID: PMC3167626
- DOI: 10.1093/nar/gkr504
Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods
Abstract
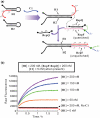
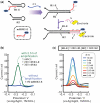
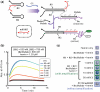
Signal amplification is a key component of molecular detection. Enzyme-free signal amplification is especially appealing for the development of low-cost, point-of-care diagnostics. It has been previously shown that enzyme-free DNA circuits with signal-amplification capacity can be designed using a mechanism called 'catalyzed hairpin assembly'. However, it is unclear whether the efficiency and modularity of such circuits is suitable for multiple analytical applications. We have therefore designed and characterized a simplified DNA circuit based on catalyzed hairpin assembly, and applied it to multiple different analytical formats, including fluorescent, colorimetric, and electrochemical and signaling. By optimizing the design of previous hairpin-based catalytic assemblies we found that our circuit has almost zero background and a high catalytic efficiency, with a k(cat) value above 1 min(-1). The inherent modularity of the circuit allowed us to readily adapt our circuit to detect both RNA and small molecule analytes. Overall, these data demonstrate that catalyzed hairpin assembly is suitable for analyte detection and signal amplification in a 'plug-and-play' fashion.
Figures



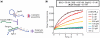

References
-
- Adler M, Wacker R, Niemeyer CM. Sensitivity by combination: immuno-PCR and related technologies. Analyst. 2008;133:702–718. - PubMed
-
- Niemeyer CM, Adler M, Wacker R. Detecting antigens by quantitative immuno-PCR. Nat. Protoc. 2007;2:1918–1930. - PubMed
-
- Zhao W, Ali MM, Brook MA, Li Y. Rolling circle amplification: applications in nanotechnology and biodetection with functional nucleic acids. Angew. Chem. Int. Ed. Engl. 2008;47:6330–6337. - PubMed
-
- Cho EJ, Yang L, Levy M, Ellington AD. Using a deoxyribozyme ligase and rolling circle amplification to detect a non-nucleic acid analyte, ATP. J. Am. Chem. Soc. 2005;127:2022–2023. - PubMed
-
- Gullberg M, Fredriksson S, Taussig M, Jarvius J, Gustafsdottir S, Landegren U. A sense of closeness: protein detection by proximity ligation. Curr. Opin. Biotechnol. 2003;14:82–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

