Predicting selective drug targets in cancer through metabolic networks
- PMID: 21694718
- PMCID: PMC3159974
- DOI: 10.1038/msb.2011.35
Predicting selective drug targets in cancer through metabolic networks
Erratum in
- Mol Syst Biol. 2011;7. doi:10.1038/msb.2011.51
Abstract
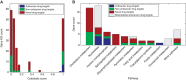
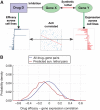
The interest in studying metabolic alterations in cancer and their potential role as novel targets for therapy has been rejuvenated in recent years. Here, we report the development of the first genome-scale network model of cancer metabolism, validated by correctly identifying genes essential for cellular proliferation in cancer cell lines. The model predicts 52 cytostatic drug targets, of which 40% are targeted by known, approved or experimental anticancer drugs, and the rest are new. It further predicts combinations of synthetic lethal drug targets, whose synergy is validated using available drug efficacy and gene expression measurements across the NCI-60 cancer cell line collection. Finally, potential selective treatments for specific cancers that depend on cancer type-specific downregulation of gene expression and somatic mutations are compiled.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Comment in
-
Systems biology: Lethal weaknesses.Nat Rev Cancer. 2011 Jul 7;11(8):538-9. doi: 10.1038/nrc3109. Nat Rev Cancer. 2011. PMID: 21734723 No abstract available.
Similar articles
-
Predicting drug targets and biomarkers of cancer via genome-scale metabolic modeling.Clin Cancer Res. 2012 Oct 15;18(20):5572-84. doi: 10.1158/1078-0432.CCR-12-1856. Clin Cancer Res. 2012. PMID: 23071359 Review.
-
Malignancy of Cancers and Synthetic Lethal Interactions Associated With Mutations of Cancer Driver Genes.Medicine (Baltimore). 2016 Feb;95(8):e2697. doi: 10.1097/MD.0000000000002697. Medicine (Baltimore). 2016. PMID: 26937901 Free PMC article.
-
Identification of selective cytotoxic and synthetic lethal drug responses in triple negative breast cancer cells.Mol Cancer. 2016 May 10;15(1):34. doi: 10.1186/s12943-016-0517-3. Mol Cancer. 2016. PMID: 27165605 Free PMC article.
-
Controllability in cancer metabolic networks according to drug targets as driver nodes.PLoS One. 2013 Nov 25;8(11):e79397. doi: 10.1371/journal.pone.0079397. eCollection 2013. PLoS One. 2013. PMID: 24282504 Free PMC article.
-
Modeling cancer metabolism on a genome scale.Mol Syst Biol. 2015 Jun 30;11(6):817. doi: 10.15252/msb.20145307. Mol Syst Biol. 2015. PMID: 26130389 Free PMC article. Review.
Cited by
-
Breast cancer dormancy: need for clinically relevant models to address current gaps in knowledge.NPJ Breast Cancer. 2021 May 28;7(1):66. doi: 10.1038/s41523-021-00269-x. NPJ Breast Cancer. 2021. PMID: 34050189 Free PMC article. Review.
-
Flux balance analysis predicts essential genes in clear cell renal cell carcinoma metabolism.Sci Rep. 2015 Jun 4;5:10738. doi: 10.1038/srep10738. Sci Rep. 2015. PMID: 26040780 Free PMC article.
-
Ensemble modeling of cancer metabolism.Front Physiol. 2012 May 16;3:135. doi: 10.3389/fphys.2012.00135. eCollection 2012. Front Physiol. 2012. PMID: 22623918 Free PMC article.
-
The dichotomy in degree correlation of biological networks.PLoS One. 2011;6(12):e28322. doi: 10.1371/journal.pone.0028322. Epub 2011 Dec 2. PLoS One. 2011. PMID: 22164269 Free PMC article.
-
A network inference method for large-scale unsupervised identification of novel drug-drug interactions.PLoS Comput Biol. 2013;9(12):e1003374. doi: 10.1371/journal.pcbi.1003374. Epub 2013 Dec 5. PLoS Comput Biol. 2013. PMID: 24339767 Free PMC article.
References
-
- Arikketh D, Nelson R, Vance JE (2008) Defining the importance of phosphatidylserine synthase-1 (PSS1): unexpected viability of PSS1-deficient mice. J Biol Chem 283: 12888–12897 - PubMed
-
- Berenbaum MC (1989) What is synergy? Pharmacol Rev 41: 93–141 - PubMed
-
- Bichsel VE, Liotta LA, Petricoin EF III (2001) Cancer proteomics: from biomarker discovery to signal pathway profiling. Cancer J 7: 69–78 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

