Autoregulation of Parkin activity through its ubiquitin-like domain
- PMID: 21694720
- PMCID: PMC3160258
- DOI: 10.1038/emboj.2011.204
Autoregulation of Parkin activity through its ubiquitin-like domain
Abstract
Parkin is an E3-ubiquitin ligase belonging to the RBR (RING-InBetweenRING-RING family), and is involved in the neurodegenerative disorder Parkinson's disease. Autosomal recessive juvenile Parkinsonism, which is one of the most common familial forms of the disease, is directly linked to mutations in the parkin gene. However, the molecular mechanisms of Parkin dysfunction in the disease state remain to be established. We now demonstrate that the ubiquitin-like domain of Parkin functions to inhibit its autoubiquitination. Moreover pathogenic Parkin mutations disrupt this autoinhibition, resulting in a constitutively active molecule. In addition, we show that the mechanism of autoregulation involves ubiquitin binding by a C-terminal region of Parkin. Our observations provide important molecular insights into the underlying basis of Parkinson's disease, and in the regulation of RBR E3-ligase activity.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



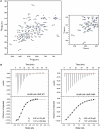


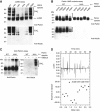

Comment in
-
Policing Parkin with a UblD.EMBO J. 2011 Jul 20;30(14):2757-8. doi: 10.1038/emboj.2011.223. EMBO J. 2011. PMID: 21772326 Free PMC article.
References
-
- Biasini E, Fioriti L, Ceglia I, Invernizzi R, Bertoli A, Chiesa R, Forloni G (2004) Proteasome inhibition and aggregation in Parkinson’s disease: a comparative study in untransfected and transfected cells. J Neurochem 88: 545–553 - PubMed
-
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 - PubMed
-
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM (2004) S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science 304: 1328–1331 - PubMed
-
- Cooper HJ, Heath JK, Jaffray E, Hay RT, Lam TT, Marshall AG (2004) Identification of sites of ubiquitination in proteins: a fourier transform ion cyclotron resonance mass spectrometry approach. Anal Chem 76: 6982–6988 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

