Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription
- PMID: 21694722
- PMCID: PMC3160253
- DOI: 10.1038/emboj.2011.194
Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription
Abstract
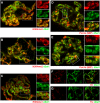
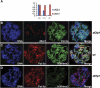
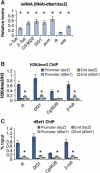
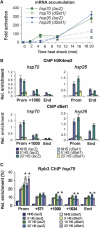
Histone H3 lysine 4 trimethylation (H3K4me3) is a major hallmark of promoter-proximal histones at transcribed genes. Here, we report that a previously uncharacterized Drosophila H3K4 methyltransferase, dSet1, and not the other putative histone H3K4 methyltransferases (Trithorax; Trithorax-related protein), is predominantly responsible for histone H3K4 trimethylation. Functional and proteomics studies reveal that dSet1 is a component of a conserved H3K4 trimethyltransferase complex and polytene staining and live cell imaging assays show widespread association of dSet1 with transcriptionally active genes. dSet1 is present at the promoter region of all tested genes, including activated Hsp70 and Hsp26 heat shock genes and is required for optimal mRNA accumulation from the tested genes. In the case of Hsp70, the mRNA production defect in dSet1 RNAi-treated cells is accompanied by retention of Pol II at promoters. Our data suggest that dSet1-dependent H3K4me3 is responsible for the generation of a chromatin structure at active promoters that ensures optimal Pol II release into productive elongation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Allis CD, Berger SL, Cote J, Dent S, Jenuwein T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y (2007) New nomenclature for chromatin-modifying enzymes. Cell 131: 633–636 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

