Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules
- PMID: 21694767
- PMCID: PMC3111414
- DOI: 10.1371/journal.pone.0016907
Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules
Abstract
Background: Circadian clocks provide an adaptive advantage through anticipation of daily and seasonal environmental changes. In plants, the central clock oscillator is regulated by several interlocking feedback loops. It was shown that a substantial proportion of the Arabidopsis genome cycles with phases of peak expression covering the entire day. Synchronized transcriptome cycling is driven through an extensive network of diurnal and clock-regulated transcription factors and their target cis-regulatory elements. Study of the cycling transcriptome in other plant species could thus help elucidate the similarities and differences and identify hubs of regulation common to monocot and dicot plants.
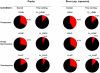
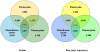
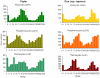
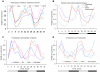
Methodology/principal findings: Using a combination of oligonucleotide microarrays and data mining pipelines, we examined daily rhythms in gene expression in one monocotyledonous and one dicotyledonous plant, rice and poplar, respectively. Cycling transcriptomes were interrogated under different diurnal (driven) and circadian (free running) light and temperature conditions. Collectively, photocycles and thermocycles regulated about 60% of the expressed nuclear genes in rice and poplar. Depending on the condition tested, up to one third of oscillating Arabidopsis-poplar-rice orthologs were phased within three hours of each other suggesting a high degree of conservation in terms of rhythmic gene expression. We identified clusters of rhythmically co-expressed genes and searched their promoter sequences to identify phase-specific cis-elements, including elements that were conserved in the promoters of Arabidopsis, poplar, and rice.
Conclusions/significance: Our results show that the cycling patterns of many circadian clock genes are highly conserved across poplar, rice, and Arabidopsis. The expression of many orthologous genes in key metabolic and regulatory pathways is diurnal and/or circadian regulated and phased to similar times of day. Our results confirm previous findings in Arabidopsis of three major classes of cis-regulatory modules within the plant circadian network: the morning (ME, GBOX), evening (EE, GATA), and midnight (PBX/TBX/SBX) modules. Identification of identical overrepresented motifs in the promoters of cycling genes from different species suggests that the core diurnal/circadian cis-regulatory network is deeply conserved between mono- and dicotyledonous species.
Conflict of interest statement
Figures





References
-
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

