A second endolysin gene is fully embedded in-frame with the lysA gene of mycobacteriophage Ms6
- PMID: 21694774
- PMCID: PMC3111421
- DOI: 10.1371/journal.pone.0020515
A second endolysin gene is fully embedded in-frame with the lysA gene of mycobacteriophage Ms6
Abstract
Mycobacteriophages are dsDNA viruses that infect mycobacterial hosts. The mycobacteriophage Ms6 accomplishes lysis by producing two cell wall hydrolytic enzymes, Lysin A (LysA) that possesses a central peptidoglycan recognition protein (PGRP) super-family conserved domain with the amidase catalytic site, that cleaves the amide bond between the N-acetylmuramic acid and L-alanine residues in the oligopeptide crosslinking chains of the peptidoglycan and Lysin B (LysB) a mycolylarabinogalactan esterase that hydrolyzes the mycolic acids from the mycolyl-arabinogalactan-peptidoglycan complex. Examination of the endolysin (lysA) DNA sequence revealed the existence of an embedded gene (lysA(241)) encoded in the same reading frame and preceded by a consensus ribosome-binding site. In the present work we show that, even though lysA is essential for Ms6 viability, phage mutants that express only the longer (Lysin(384)) or the shorter (Lysin(241)) endolysin are viable, but defective in the normal timing, progression and completion of host cell lysis. In addition, both endolysins have peptidoglycan hydrolase activity and demonstrated broad growth inhibition activity against various gram-positive bacteria and mycobacteria.
Conflict of interest statement
Figures
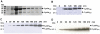
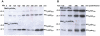



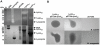
References
-
- Young R, Wang IN, Roof WD. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–127. - PubMed
-
- Young R. Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol. 2002;4:21–36. - PubMed
-
- São-José C, Nascimento J, Parreira R, Santos M. Release of progeny phages from infected cells. In: McGrath S, van Sinderen D, editors. Bacteriophage: genetics and molecular biology. Caister Academic Press; 2007. pp. 309–336.
-
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

