The metabolic serine hydrolases and their functions in mammalian physiology and disease
- PMID: 21696217
- PMCID: PMC3192302
- DOI: 10.1021/cr200075y
The metabolic serine hydrolases and their functions in mammalian physiology and disease
Figures

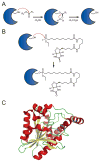
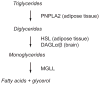
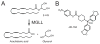

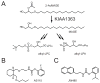

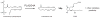

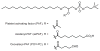
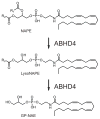




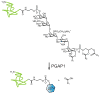
 SH (4′-phosphopantetheine). MAT* and TE* indicate that these two are SH domains. Following attachment of the initial acetate, the cycle is repeated seven times to generate one equivalent of palmitic acid.
SH (4′-phosphopantetheine). MAT* and TE* indicate that these two are SH domains. Following attachment of the initial acetate, the cycle is repeated seven times to generate one equivalent of palmitic acid.






References
-
- Powers JC, Asgian JL, Ekici OD, James KE. Chem Rev. 2002;102:4639. - PubMed
-
- Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. Academic Press; Bath: 1998.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

