Fusion of metabolomics and proteomics data for biomarkers discovery: case study on the experimental autoimmune encephalomyelitis
- PMID: 21696593
- PMCID: PMC3225201
- DOI: 10.1186/1471-2105-12-254
Fusion of metabolomics and proteomics data for biomarkers discovery: case study on the experimental autoimmune encephalomyelitis
Abstract
Background: Analysis of Cerebrospinal Fluid (CSF) samples holds great promise to diagnose neurological pathologies and gain insight into the molecular background of these pathologies. Proteomics and metabolomics methods provide invaluable information on the biomolecular content of CSF and thereby on the possible status of the central nervous system, including neurological pathologies. The combined information provides a more complete description of CSF content. Extracting the full combined information requires a combined analysis of different datasets i.e. fusion of the data.
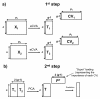
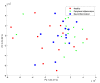
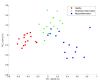
Results: A novel fusion method is presented and applied to proteomics and metabolomics data from a pre-clinical model of multiple sclerosis: an Experimental Autoimmune Encephalomyelitis (EAE) model in rats. The method follows a mid-level fusion architecture. The relevant information is extracted per platform using extended canonical variates analysis. The results are subsequently merged in order to be analyzed jointly. We find that the combined proteome and metabolome data allow for the efficient and reliable discrimination between healthy, peripherally inflamed rats, and rats at the onset of the EAE. The predicted accuracy reaches 89% on a test set. The important variables (metabolites and proteins) in this model are known to be linked to EAE and/or multiple sclerosis.
Conclusions: Fusion of proteomics and metabolomics data is possible. The main issues of high-dimensionality and missing values are overcome. The outcome leads to higher accuracy in prediction and more exhaustive description of the disease profile. The biological interpretation of the involved variables validates our fusion approach.
Figures


Similar articles
-
Cerebrospinal Fluid Metabolomics and Proteomics Integration in Neurological Syndromes.Methods Mol Biol. 2025;2914:303-321. doi: 10.1007/978-1-0716-4462-1_21. Methods Mol Biol. 2025. PMID: 40167926 Review.
-
Simultaneous analysis of plasma and CSF by NMR and hierarchical models fusion.Anal Bioanal Chem. 2012 May;403(4):947-59. doi: 10.1007/s00216-012-5871-4. Epub 2012 Mar 7. Anal Bioanal Chem. 2012. PMID: 22395451 Free PMC article.
-
Interpretation and visualization of non-linear data fusion in kernel space: study on metabolomic characterization of progression of multiple sclerosis.PLoS One. 2012;7(6):e38163. doi: 10.1371/journal.pone.0038163. Epub 2012 Jun 8. PLoS One. 2012. PMID: 22715376 Free PMC article. Clinical Trial.
-
Metabolomics approaches in experimental allergic encephalomyelitis.J Neuroimmunol. 2018 Jan 15;314:94-100. doi: 10.1016/j.jneuroim.2017.11.018. Epub 2017 Dec 2. J Neuroimmunol. 2018. PMID: 29224959
-
Ten years of proteomics in multiple sclerosis.Proteomics. 2014 Mar;14(4-5):467-80. doi: 10.1002/pmic.201300268. Proteomics. 2014. PMID: 24339438 Review.
Cited by
-
Cerebrospinal Fluid Metabolomics and Proteomics Integration in Neurological Syndromes.Methods Mol Biol. 2025;2914:303-321. doi: 10.1007/978-1-0716-4462-1_21. Methods Mol Biol. 2025. PMID: 40167926 Review.
-
MicroRNA-205 targets tight junction-related proteins during urothelial cellular differentiation.Mol Cell Proteomics. 2014 Sep;13(9):2321-36. doi: 10.1074/mcp.M113.033563. Epub 2014 Jun 9. Mol Cell Proteomics. 2014. PMID: 24912853 Free PMC article.
-
Predicting changes in renal metabolism after compound exposure with a genome-scale metabolic model.Toxicol Appl Pharmacol. 2021 Feb 1;412:115390. doi: 10.1016/j.taap.2020.115390. Epub 2020 Dec 31. Toxicol Appl Pharmacol. 2021. PMID: 33387578 Free PMC article.
-
Metabolomics as a promising tool for improving understanding of multiple sclerosis: A review of recent advances.Biomed J. 2022 Aug;45(4):594-606. doi: 10.1016/j.bj.2022.01.004. Epub 2022 Jan 15. Biomed J. 2022. PMID: 35042018 Free PMC article. Review.
-
Cerebrospinal fluid metabolomics: detection of neuroinflammation in human central nervous system disease.Clin Transl Immunology. 2021 Aug 6;10(8):e1318. doi: 10.1002/cti2.1318. eCollection 2021. Clin Transl Immunology. 2021. PMID: 34386234 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

