Campylobacter jejuni group III phage CP81 contains many T4-like genes without belonging to the T4-type phage group: implications for the evolution of T4 phages
- PMID: 21697478
- PMCID: PMC3165843
- DOI: 10.1128/JVI.00395-11
Campylobacter jejuni group III phage CP81 contains many T4-like genes without belonging to the T4-type phage group: implications for the evolution of T4 phages
Abstract
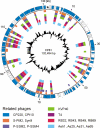
CP81 is a virulent Campylobacter group III phage whose linear genome comprises 132,454 bp. At the nucleotide level, CP81 differs from other phages. However, a number of its structural and replication/recombination proteins revealed a relationship to the group II Campylobacter phages CP220/CPt10 and to T4-type phages. Unlike the T4-related phages, the CP81 genome does not contain conserved replication and virion modules. Instead, the respective genes are scattered throughout the phage genome. Moreover, most genes for metabolic enzymes of CP220/CPt10 are lacking in CP81. On the other hand, the CP81 genome contains nine similar genes for homing endonucleases which may be involved in the attrition of the conserved gene order for the virion core genes of T4-type phages. The phage apparently possesses an unusual modification of C or G bases. Efficient cleavage of its DNA was only achieved with restriction enzymes recognizing pure A/T sites. Uncommonly, phenol extraction leads to a significant loss of CP81 DNA from the aqueous layer, a property not yet described for other phages belonging to the T4 superfamily.
Figures





Similar articles
-
Genome and proteome of Campylobacter jejuni bacteriophage NCTC 12673.Appl Environ Microbiol. 2011 Dec;77(23):8265-71. doi: 10.1128/AEM.05562-11. Epub 2011 Sep 30. Appl Environ Microbiol. 2011. PMID: 21965409 Free PMC article.
-
The genome and proteome of a Campylobacter coli bacteriophage vB_CcoM-IBB_35 reveal unusual features.Virol J. 2012 Jan 27;9:35. doi: 10.1186/1743-422X-9-35. Virol J. 2012. PMID: 22284308 Free PMC article.
-
Primary isolation strain determines both phage type and receptors recognised by Campylobacter jejuni bacteriophages.PLoS One. 2015 Jan 13;10(1):e0116287. doi: 10.1371/journal.pone.0116287. eCollection 2015. PLoS One. 2015. PMID: 25585385 Free PMC article.
-
The SPO1-related bacteriophages.Arch Virol. 2010 Oct;155(10):1547-61. doi: 10.1007/s00705-010-0783-0. Epub 2010 Aug 17. Arch Virol. 2010. PMID: 20714761 Review.
-
Evolution of T4-related phages.Virus Genes. 1995;11(2-3):285-97. doi: 10.1007/BF01728666. Virus Genes. 1995. PMID: 8828153 Review.
Cited by
-
The complete genome sequence of bacteriophage CP21 reveals modular shuffling in Campylobacter group II phages.J Virol. 2012 Aug;86(16):8896. doi: 10.1128/JVI.01252-12. J Virol. 2012. PMID: 22843857 Free PMC article.
-
Binding Specificities of the Telomere Phage ϕKO2 Prophage Repressor CB and Lytic Repressor Cro.Viruses. 2016 Aug 3;8(8):213. doi: 10.3390/v8080213. Viruses. 2016. PMID: 27527206 Free PMC article.
-
Campylobacter Phage Isolation and Characterization: What We Have Learned So Far.Methods Protoc. 2019 Feb 15;2(1):18. doi: 10.3390/mps2010018. Methods Protoc. 2019. PMID: 31164600 Free PMC article. Review.
-
Application of Campylobacter jejuni Phages: Challenges and Perspectives.Animals (Basel). 2020 Feb 11;10(2):279. doi: 10.3390/ani10020279. Animals (Basel). 2020. PMID: 32054081 Free PMC article. Review.
-
Analysis of the First Temperate Broad Host Range Brucellaphage (BiPBO1) Isolated from B. inopinata.Front Microbiol. 2016 Jan 28;7:24. doi: 10.3389/fmicb.2016.00024. eCollection 2016. Front Microbiol. 2016. PMID: 26858702 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

