Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication
- PMID: 21697487
- PMCID: PMC3165839
- DOI: 10.1128/JVI.00059-11
Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication
Abstract
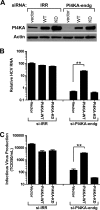
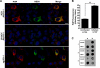
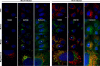
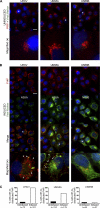
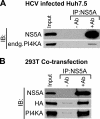
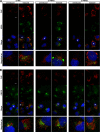
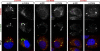
Phosphatidylinositol 4-kinase III alpha (PI4KA) is an essential cofactor of hepatitis C virus (HCV) replication. We initiated this study to determine whether HCV directly engages PI4KA to establish its replication. PI4KA kinase activity was found to be absolutely required for HCV replication using a small interfering RNA transcomplementation assay. Moreover, HCV infection or subgenomic HCV replicons produced a dramatic increase in phosphatidylinositol 4-phosphate (PI4P) accumulation throughout the cytoplasm, which partially colocalized with the endoplasmic reticulum. In contrast, the majority of PI4P accumulated at the Golgi bodies in uninfected cells. The increase in PI4P was not observed after infection with UV-inactivated HCV and did not reflect changes in PI4KA protein or RNA abundance. In an analysis of U2OS cell lines with inducible expression of the HCV polyprotein or individual viral proteins, viral polyprotein expression resulted in enhanced cytoplasmic PI4P production. Increased PI4P accumulation following HCV protein expression was precluded by silencing the expression of PI4KA, but not the related PI4KB. Silencing PI4KA also resulted in aberrant agglomeration of viral replicase proteins, including NS5A, NS5B, and NS3. NS5A alone, but not other viral proteins, stimulated PI4P production in vivo and enhanced PI4KA kinase activity in vitro. Lastly, PI4KA coimmunoprecipitated with NS5A from infected Huh-7.5 cells and from dually transfected 293T cells. In sum, these results suggest that HCV NS5A modulation of PI4KA-dependent PI4P production influences replication complex formation.
Figures



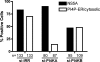
References
-
- Ahn J., et al. 2004. Systematic identification of hepatocellular proteins interacting with NS5A of the hepatitis C virus. J. Biochem. Mol. Biol. 37:741–748 - PubMed
-
- Balla A., Balla T. 2006. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 16:351–361 - PubMed
-
- Balla A., Tuymetova G., Barshishat M., Geiszt M., Balla T. 2002. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J. Biol. Chem. 277:20041–20050 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

