A general strategy for the evolution of bond-forming enzymes using yeast display
- PMID: 21697512
- PMCID: PMC3136257
- DOI: 10.1073/pnas.1101046108
A general strategy for the evolution of bond-forming enzymes using yeast display
Abstract
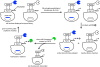
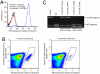
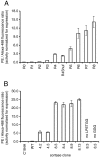
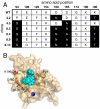
The ability to routinely generate efficient protein catalysts of bond-forming reactions chosen by researchers, rather than nature, is a long-standing goal of the molecular life sciences. Here, we describe a directed evolution strategy for enzymes that catalyze, in principle, any bond-forming reaction. The system integrates yeast display, enzyme-mediated bioconjugation, and fluorescence-activated cell sorting to isolate cells expressing proteins that catalyze the coupling of two substrates chosen by the researcher. We validated the system using model screens for Staphylococcus aureus sortase A-catalyzed transpeptidation activity, resulting in enrichment factors of 6,000-fold after a single round of screening. We applied the system to evolve sortase A for improved catalytic activity. After eight rounds of screening, we isolated variants of sortase A with up to a 140-fold increase in LPETG-coupling activity compared with the starting wild-type enzyme. An evolved sortase variant enabled much more efficient labeling of LPETG-tagged human CD154 expressed on the surface of HeLa cells compared with wild-type sortase. Because the method developed here does not rely on any particular screenable or selectable property of the substrates or product, it represents a powerful alternative to existing enzyme evolution methods.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Savile CK, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329:305–309. - PubMed
-
- Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: A versatile method for protein labeling. Nat Chem Biol. 2007;3:707–708. - PubMed
-
- Walsh G. Biopharmaceutical benchmarks 2006. Nat Biotechnol. 2006;24:769–776. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

