Analysis of bacterial core communities in the central Baltic by comparative RNA-DNA-based fingerprinting provides links to structure-function relationships
- PMID: 21697960
- PMCID: PMC3246236
- DOI: 10.1038/ismej.2011.80
Analysis of bacterial core communities in the central Baltic by comparative RNA-DNA-based fingerprinting provides links to structure-function relationships
Abstract
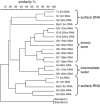
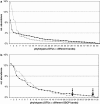
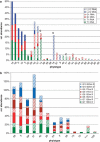
Understanding structure-function links of microbial communities is a central theme of microbial ecology since its beginning. To this end, we studied the spatial variability of the bacterioplankton community structure and composition across the central Baltic Sea at four stations, which were up to 450 km apart and at a depth profile representative for the central part (Gotland Deep, 235 m). Bacterial community structure was followed by 16S ribosomal RNA (rRNA)- and 16S rRNA gene-based fingerprints using single-strand conformation polymorphism (SSCP) electrophoresis. Species composition was determined by sequence analysis of SSCP bands. High similarities of the bacterioplankton communities across several hundred kilometers were observed in the surface water using RNA- and DNA-based fingerprints. In these surface communities, the RNA- and DNA-based fingerprints resulted in very different pattern, presumably indicating large difference between the active members of the community as represented by RNA-based fingerprints and the present members represented by the DNA-based fingerprints. This large discrepancy changed gradually over depth, resulting in highly similar RNA- and DNA-based fingerprints in the anoxic part of the water column below 130 m depth. A conceivable mechanism explaining this high similarity could be the reduced oxidative stress in the anoxic zone. The stable communities on the surface and in the anoxic zone indicate the strong influence of the hydrography on the bacterioplankton community structure. Comparative analysis of RNA- and DNA-based community structure provided criteria for the identification of the core community, its key members and their links to biogeochemical functions.
Figures

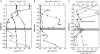


References
-
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2009;4:171–181. - PubMed
-
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5:782–791. - PubMed
-
- Bassam BJ, Caetano-Anolles G, Gresshoff PM. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;80:81–84. - PubMed

