Variation in human recombination rates and its genetic determinants
- PMID: 21698098
- PMCID: PMC3117798
- DOI: 10.1371/journal.pone.0020321
Variation in human recombination rates and its genetic determinants
Abstract
Background: Despite the fundamental role of crossing-over in the pairing and segregation of chromosomes during human meiosis, the rates and placements of events vary markedly among individuals. Characterizing this variation and identifying its determinants are essential steps in our understanding of the human recombination process and its evolution.
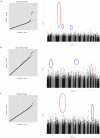
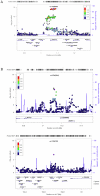
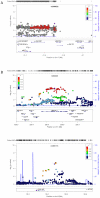
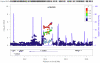
Study design/results: Using three large sets of European-American pedigrees, we examined variation in five recombination phenotypes that capture distinct aspects of crossing-over patterns. We found that the mean recombination rate in males and females and the historical hotspot usage are significantly heritable and are uncorrelated with one another. We then conducted a genome-wide association study in order to identify loci that influence them. We replicated associations of RNF212 with the mean rate in males and in females as well as the association of Inversion 17q21.31 with the female mean rate. We also replicated the association of PRDM9 with historical hotspot usage, finding that it explains most of the genetic variance in this phenotype. In addition, we identified a set of new candidate regions for further validation.
Significance: These findings suggest that variation at broad and fine scales is largely separable and that, beyond three known loci, there is no evidence for common variation with large effects on recombination phenotypes.
Conflict of interest statement
Figures


References
-
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. - PubMed
-
- Sturtevant AH. The behavior of the chromosome as studied through linkage. Molecular and General Genetics MGG. 1915;13:234–287.
-
- Muller HJ. The mechanism of crossing-over. American Naturalist. 1916;50:421–434.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

