Collective dynamics of gene expression in cell populations
- PMID: 21698278
- PMCID: PMC3115940
- DOI: 10.1371/journal.pone.0020530
Collective dynamics of gene expression in cell populations
Abstract
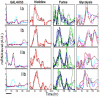
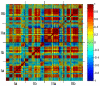
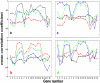
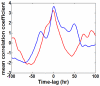
The phenotypic state of the cell is commonly thought to be determined by the set of expressed genes. However, given the apparent complexity of genetic networks, it remains open what processes stabilize a particular phenotypic state. Moreover, it is not clear how unique is the mapping between the vector of expressed genes and the cell's phenotypic state. To gain insight on these issues, we study here the expression dynamics of metabolically essential genes in twin cell populations. We show that two yeast cell populations derived from a single steady-state mother population and exhibiting a similar growth phenotype in response to an environmental challenge, displayed diverse expression patterns of essential genes. The observed diversity in the mean expression between populations could not result from stochastic cell-to-cell variability, which would be averaged out in our large cell populations. Remarkably, within a population, sets of expressed genes exhibited coherent dynamics over many generations. Thus, the emerging gene expression patterns resulted from collective population dynamics. It suggests that in a wide range of biological contexts, gene expression reflects a self-organization process coupled to population-environment dynamics.
Conflict of interest statement
Figures





References
-
- Booth IR. Stress and the single cell: intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Food Microbiology. 2002;71:19–30. - PubMed
-
- Kærn M, Elston TC, Blake WJ, Collins JJ. 2005;6:451–464. Stochasticity in gene expression: from theories to phenotypes. - PubMed
-
- Brenner N, Farkash K, Braun E. Dynamics of protein distributions in cell populations. Physical Biology. 2006;3:172–182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

