Pharmacological modulation of chemokine receptor function
- PMID: 21699506
- PMCID: PMC3372818
- DOI: 10.1111/j.1476-5381.2011.01551.x
Pharmacological modulation of chemokine receptor function
Abstract
G protein-coupled chemokine receptors and their peptidergic ligands are interesting therapeutic targets due to their involvement in various immune-related diseases, including rheumatoid arthritis, multiple sclerosis, inflammatory bowel disease, chronic obstructive pulmonary disease, HIV-1 infection and cancer. To tackle these diseases, a lot of effort has been focused on discovery and development of small-molecule chemokine receptor antagonists. This has been rewarded by the market approval of two novel chemokine receptor inhibitors, AMD3100 (CXCR4) and Maraviroc (CCR5) for stem cell mobilization and treatment of HIV-1 infection respectively. The recent GPCR crystal structures together with mutagenesis and pharmacological studies have aided in understanding how small-molecule ligands interact with chemokine receptors. Many of these ligands display behaviour deviating from simple competition and do not interact with the chemokine binding site, providing evidence for an allosteric mode of action. This review aims to give an overview of the evidence supporting modulation of this intriguing receptor family by a range of ligands, including small molecules, peptides and antibodies. Moreover, the computer-assisted modelling of chemokine receptor-ligand interactions is discussed in view of GPCR crystal structures. Finally, the implications of concepts such as functional selectivity and chemokine receptor dimerization are considered.
© 2011 The Authors. British Journal of Pharmacology © 2011 The British Pharmacological Society.
Figures

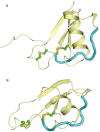

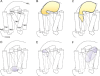


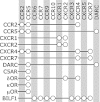
References
-
- Ahuja S, Lee J, Murphy P. CXC chemokines bind to unique sets of selectivity determinants that can function independently and are broadly distributed on multiple domains of human interleukin-8 receptor B. Determinants of high affinity binding and receptor activation are distinct. J Biol Chem. 1996;271:225–232. - PubMed
-
- Allegretti M, Bertini R, Cesta M, Bizzarri C, Di Bitondo R, Di Cioccio V, et al. 2-Arylpropionic CXC chemokine receptor 1 (CXCR1) ligands as novel noncompetitive CXCL8 inhibitors. J Med Chem. 2005;48:4312–4331. - PubMed
-
- Allen S, Crown S, Handel T. Chemokine: receptor structure, interactions, and antagonism. Annu Rev Immunol. 2007;25:787–820. - PubMed
-
- Andrews G, Jones C, Wreggett KA. An intracellular allosteric site for a specific class of antagonists of the CC chemokine G protein-coupled receptors CCR4 and CCR5. Mol Pharmacol. 2008;73:855–867. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

