Variations on a theme: diversification of cuticular hydrocarbons in a clade of cactophilic Drosophila
- PMID: 21699713
- PMCID: PMC3161901
- DOI: 10.1186/1471-2148-11-179
Variations on a theme: diversification of cuticular hydrocarbons in a clade of cactophilic Drosophila
Abstract
Background: We characterized variation and chemical composition of epicuticular hydrocarbons (CHCs) in the seven species of the Drosophila buzzatii cluster with gas chromatography/mass spectrometry. Despite the critical role of CHCs in providing resistance to desiccation and involvement in communication, such as courtship behavior, mating, and aggregation, few studies have investigated how CHC profiles evolve within and between species in a phylogenetic context. We analyzed quantitative differences in CHC profiles in populations of the D. buzzatii species cluster in order to assess the concordance of CHC differentiation with species divergence.
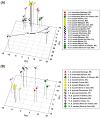
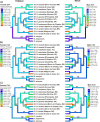
Results: Thirty-six CHC components were scored in single fly extracts with carbon chain lengths ranging from C29 to C39, including methyl-branched alkanes, n-alkenes, and alkadienes. Multivariate analysis of variance revealed that CHC amounts were significantly different among all species and canonical discriminant function (CDF) analysis resolved all species into distinct, non-overlapping groups. Significant intraspecific variation was found in different populations of D. serido suggesting that this taxon is comprised of at least two species. We summarized CHC variation using CDF analysis and mapped the first five CHC canonical variates (CVs) onto an independently derived period (per) gene + chromosome inversion + mtDNA COI gene for each sex. We found that the COI sequences were not phylogenetically informative due to introgression between some species, so only per + inversion data were used. Positive phylogenetic signal was observed mainly for CV1 when parsimony methods and the test for serial independence (TFSI) were used. These results changed when no outgroup species were included in the analysis and phylogenetic signal was then observed for female CV3 and/or CV4 and male CV4 and CV5. Finally, removal of divergent populations of D. serido significantly increased the amount of phylogenetic signal as up to four out of five CVs then displayed positive phylogenetic signal.
Conclusions: CHCs were conserved among species while quantitative differences in CHC profiles between populations and species were statistically significant. Most CHCs were species-, population-, and sex-specific. Mapping CHCs onto an independently derived phylogeny revealed that a significant portion of CHC variation was explained by species' systematic affinities indicating phylogenetic conservatism in the evolution of these hydrocarbon arrays, presumptive waterproofing compounds and courtship signals as in many other drosophilid species.
Figures




References
-
- Harvey PH, Pagel MD. The comparative method in evolutionary biology. New York: Oxford Univ. Press; 1991.
-
- Blomberg SP, Garland T Jr, Ives AR. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution. 2003;57(4):717–745. - PubMed
-
- Wimberger PH, de Queiroz A. In: Phylogenies and the Comparative Method in Animal Behavior. Martins EP, editor. New York: Oxford University Press; 1996. Comparing behavioral and morphological characters as indicators of phylogeny; pp. 206–233.
-
- Duckworth RA. The role of behavior in evolution: a search for mechanism. Evol Ecol. 2009;23:513–531. doi: 10.1007/s10682-008-9252-6. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

