Development of small molecule inhibitors and probes of human SUMO deconjugating proteases
- PMID: 21700208
- PMCID: PMC3131534
- DOI: 10.1016/j.chembiol.2011.05.008
Development of small molecule inhibitors and probes of human SUMO deconjugating proteases
Abstract
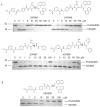
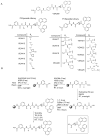
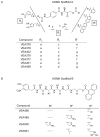
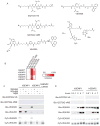
Sentrin specific proteases (SENPs) are responsible for activating and deconjugating SUMO (Small Ubiquitin like MOdifier) from target proteins. It remains difficult to study this posttranslational modification due to the lack of reagents that can be used to block the removal of SUMO from substrates. Here, we describe the identification of small molecule SENP inhibitors and active site probes containing aza-epoxide and acyloxymethyl ketone (AOMK) reactive groups. Both classes of compounds are effective inhibitors of hSENPs 1, 2, 5, and 7 while only the AOMKs efficiently inhibit hSENP6. Unlike previous reported peptide vinyl sulfones, these compounds covalently labeled the active site cysteine of multiple recombinantly expressed SENP proteases and the AOMK probe showed selective labeling of these SENPs when added to complex protein mixtures. The AOMK compound therefore represents promising new reagents to study the process of SUMO deconjugation.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
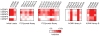

References
-
- Arastu-Kapur S, Ponder EL, Fonovic UP, Yeoh S, Yuan F, Fonovic M, Grainger M, Phillips CI, Powers JC, Bogyo M. Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat Chem Biol. 2008;4:203–213. - PubMed
-
- Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, Bogyo M. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell. 2006;23:509–521. - PubMed
-
- Borodovsky A, Ovaa H, Meester WJ, Venanzi ES, Bogyo MS, Hekking BG, Ploegh HL, Kessler BM, Overkleeft HS. Small-molecule inhibitors and probes for ubiquitin- and ubiquitin-like-specific proteases. Chembiochem. 2005;6:287–291. - PubMed
-
- Bromme D, Smith RA, Coles PJ, Kirschke H, Storer AC, Krantz A. Potent inactivation of cathepsins S and L by peptidyl (acyloxy)methyl ketones. Biol Chem Hoppe Seyler. 1994;375:343–347. - PubMed
-
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Research Materials

