Structural aspects of translation termination on the ribosome
- PMID: 21700725
- PMCID: PMC3153966
- DOI: 10.1261/rna.2733411
Structural aspects of translation termination on the ribosome
Abstract
Translation of genetic information encoded in messenger RNAs into polypeptide sequences is carried out by ribosomes in all organisms. When a full protein is synthesized, a stop codon positioned in the ribosomal A site signals termination of translation and protein release. Translation termination depends on class I release factors. Recently, atomic-resolution crystal structures were determined for bacterial 70S ribosome termination complexes bound with release factors RF1 or RF2. In combination with recent biochemical studies, the structures resolve long-standing questions about translation termination. They bring insights into the mechanisms of recognition of all three stop codons, peptidyl-tRNA hydrolysis, and coordination of stop-codon recognition with peptidyl-tRNA hydrolysis. In this review, the structural aspects of these mechanisms are discussed.
Figures

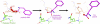
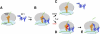
References
-
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 - PubMed
-
- Arkov AL, Murgola EJ 1999. Ribosomal RNAs in translation termination: Facts and hypotheses. Biochemistry (Mosc) 64: 1354–1359 - PubMed
-
- Baierlein C, Krebber H 2010. Translation termination: New factors and insights. RNA Biol 7: 548–550 - PubMed
-
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
