Self-recognition in social amoebae is mediated by allelic pairs of tiger genes
- PMID: 21700835
- PMCID: PMC3142563
- DOI: 10.1126/science.1203903
Self-recognition in social amoebae is mediated by allelic pairs of tiger genes
Abstract
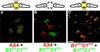
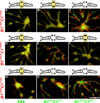
Free-living cells of the social amoebae Dictyostelium discoideum can aggregate and develop into multicellular fruiting bodies in which many die altruistically as they become stalk cells that support the surviving spores. Dictyostelium cells exhibit kin discrimination--a potential defense against cheaters, which sporulate without contributing to the stalk. Kin discrimination depends on strain relatedness, and the polymorphic genes tgrB1 and tgrC1 are potential components of that mechanism. Here, we demonstrate a direct role for these genes in kin discrimination. We show that a matching pair of tgrB1 and tgrC1 alleles is necessary and sufficient for attractive self-recognition, which is mediated by differential cell-cell adhesion. We propose that TgrB1 and TgrC1 proteins mediate this adhesion through direct binding. This system is a genetically tractable ancient model of eukaryotic self-recognition.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

