Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I
- PMID: 21700875
- PMCID: PMC3380281
- DOI: 10.1126/science.1205016
Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I
Abstract
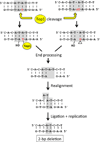
The ribonuclease (RNase) H class of enzymes degrades the RNA component of RNA:DNA hybrids and is important in nucleic acid metabolism. RNase H2 is specialized to remove single ribonucleotides [ribonucleoside monophosphates (rNMPs)] from duplex DNA, and its absence in budding yeast has been associated with the accumulation of deletions within short tandem repeats. Here, we demonstrate that rNMP-associated deletion formation requires the activity of Top1, a topoisomerase that relaxes supercoils by reversibly nicking duplex DNA. The reported studies extend the role of Top1 to include the processing of rNMPs in genomic DNA into irreversible single-strand breaks, an activity that can have distinct mutagenic consequences and may be relevant to human disease.
Figures



Comment in
-
Molecular biology. A new twist for topoisomerase.Science. 2011 Jun 24;332(6037):1510-1. doi: 10.1126/science.1208450. Science. 2011. PMID: 21700860 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

