Sleep and synaptic homeostasis: structural evidence in Drosophila
- PMID: 21700878
- PMCID: PMC3128387
- DOI: 10.1126/science.1202839
Sleep and synaptic homeostasis: structural evidence in Drosophila
Abstract
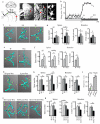
The functions of sleep remain elusive, but a strong link exists between sleep need and neuronal plasticity. We tested the hypothesis that plastic processes during wake lead to a net increase in synaptic strength and sleep is necessary for synaptic renormalization. We found that, in three Drosophila neuronal circuits, synapse size or number increases after a few hours of wake and decreases only if flies are allowed to sleep. A richer wake experience resulted in both larger synaptic growth and greater sleep need. Finally, we demonstrate that the gene Fmr1 (fragile X mental retardation 1) plays an important role in sleep-dependent synaptic renormalization.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

