MicroRNA-145 regulates human corneal epithelial differentiation
- PMID: 21701675
- PMCID: PMC3119052
- DOI: 10.1371/journal.pone.0021249
MicroRNA-145 regulates human corneal epithelial differentiation
Abstract
Background: Epigenetic factors, such as microRNAs, are important regulators in the self-renewal and differentiation of stem cells and progenies. Here we investigated the microRNAs expressed in human limbal-peripheral corneal (LPC) epithelia containing corneal epithelial progenitor cells (CEPCs) and early transit amplifying cells, and their role in corneal epithelium.
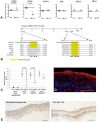
Methodology/principal findings: Human LPC epithelia was extracted for small RNAs or dissociated for CEPC culture. By Agilent Human microRNA Microarray V2 platform and GeneSpring GX11.0 analysis, we found differential expression of 18 microRNAs against central corneal (CC) epithelia, which were devoid of CEPCs. Among them, miR-184 was up-regulated in CC epithelia, similar to reported finding. Cluster miR-143/145 was expressed strongly in LPC but weakly in CC epithelia (P = 0.0004, Mann-Whitney U-test). This was validated by quantitative polymerase chain reaction (qPCR). Locked nucleic acid-based in situ hybridization on corneal rim cryosections showed miR-143/145 presence localized to the parabasal cells of limbal epithelium but negligible in basal and superficial epithelia. With holoclone forming ability, CEPCs transfected with lentiviral plasmid containing mature miR-145 sequence gave rise to defective epithelium in organotypic culture and had increased cytokeratin-3/12 and connexin-43 expressions and decreased ABCG2 and p63 compared with cells transfected with scrambled sequences. Global gene expression was analyzed using Agilent Whole Human Genome Oligo Microarray and GeneSpring GX11.0. With a 5-fold difference compared to cells with scrambled sequences, miR-145 up-regulated 324 genes (containing genes for immune response) and down-regulated 277 genes (containing genes for epithelial development and stem cell maintenance). As validated by qPCR and luciferase reporter assay, our results showed miR-145 suppressed integrin β8 (ITGB8) expression in both human corneal epithelial cells and primary CEPCs.
Conclusion/significance: We found expression of miR-143/145 cluster in human corneal epithelium. Our results also showed that miR-145 regulated the corneal epithelium formation and maintenance of epithelial integrity, via ITGB8 targeting.
Conflict of interest statement
Figures




References
-
- Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201–209. - PubMed
-
- Lehrer MS, Sun TT, Lavker RM. Strategies of epithelial repair: modulation of stem cell and transit amplifying cell proliferation. J Cell Sci. 1998;111:2867–2875. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

