RNAi-mediated inhibition of the desmosomal cadherin (desmoglein 3) impairs epithelial cell proliferation
- PMID: 21702856
- PMCID: PMC6496163
- DOI: 10.1111/j.1365-2184.2011.00765.x
RNAi-mediated inhibition of the desmosomal cadherin (desmoglein 3) impairs epithelial cell proliferation
Abstract
Objectives: Desmoglein 3 (Dsg3) is a desmosomal adhesion protein expressed in basal and immediate suprabasal layers of skin. Importance of Dsg3 in cell-cell adhesion and maintenance of tissue integrity is illustrated by findings of keratinocyte dissociation in the autoimmune disease, pemphigus vulgaris, where autoantibodies target Dsg3 on keratinocyte surfaces and cause Dsg3 depletion from desmosomes. However, recognition of possible participation of involvement of Dsg3 in cell proliferation remains controversial. Currently, available evidence suggests that Dsg3 may have both anti- and pro-proliferative roles in keratinocytes. The aim of this study was to use RNA interference (RNAi) strategy to investigate effects of silencing Dsg3 in cell-cell adhesion and cell proliferation in two cell lines, HaCaT and MDCK.
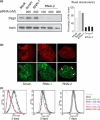
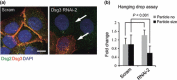
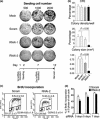
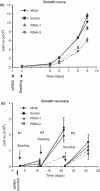
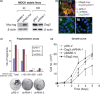
Materials and methods: Cells were transfected with siRNA, and knockdown of Dsg3 was assessed by western blotting, fluorescence-activated cell sorting and confocal microscopy. Cell-cell adhesion was analysed using the hanging drop/fragmentation assay, and cell proliferation by colony forming efficiency, BrdU incorporation, cell counts and organotypic culture.
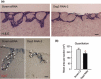
Results: Silencing Dsg3 caused defects in cell-cell adhesion and concomitant reduction in cell proliferation in both HaCaT and MDCK cells.
Conclusion: These findings suggest that Dsg3 depletion by RNAi reduces cell proliferation, which is likely to be secondary to a defect in cell-cell adhesion, an essential function required for cell differentiation and morphogenesis.
© 2011 Blackwell Publishing Ltd.
Figures
References
-
- Garrod D, Kimura TE (2008) Hyper‐adhesion: a new concept in cell‐cell adhesion. Biochem. Soc. Trans. 36, 195–201. - PubMed
-
- Sharma P, Mao X, Payne AS (2007) Beyond steric hindrance: the role of adhesion signaling pathways in the pathogenesis of pemphigus. J. Dermatol. Sci. 48, 1–14. - PubMed
-
- Garrod D, Chidgey M (2008) Desmosome structure, composition and function. Biochim. Biophys. Acta 1778, 572–587. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

