Global transcriptome response in Lactobacillus sakei during growth on ribose
- PMID: 21702908
- PMCID: PMC3146418
- DOI: 10.1186/1471-2180-11-145
Global transcriptome response in Lactobacillus sakei during growth on ribose
Abstract
Background: Lactobacillus sakei is valuable in the fermentation of meat products and exhibits properties that allow for better preservation of meat and fish. On these substrates, glucose and ribose are the main carbon sources available for growth. We used a whole-genome microarray based on the genome sequence of L. sakei strain 23K to investigate the global transcriptome response of three L. sakei strains when grown on ribose compared with glucose.
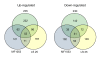
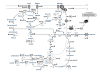
Results: The function of the common regulated genes was mostly related to carbohydrate metabolism and transport. Decreased transcription of genes encoding enzymes involved in glucose metabolism and the L-lactate dehydrogenase was observed, but most of the genes showing differential expression were up-regulated. Especially transcription of genes directly involved in ribose catabolism, the phosphoketolase pathway, and in alternative fates of pyruvate increased. Interestingly, the methylglyoxal synthase gene, which encodes an enzyme unique for L. sakei among lactobacilli, was up-regulated. Ribose catabolism seems closely linked with catabolism of nucleosides. The deoxyribonucleoside synthesis operon transcriptional regulator gene was strongly up-regulated, as well as two gene clusters involved in nucleoside catabolism. One of the clusters included a ribokinase gene. Moreover, hprK encoding the HPr kinase/phosphatase, which plays a major role in the regulation of carbon metabolism and sugar transport, was up-regulated, as were genes encoding the general PTS enzyme I and the mannose-specific enzyme II complex (EIIman). Putative catabolite-responsive element (cre) sites were found in proximity to the promoter of several genes and operons affected by the change of carbon source. This could indicate regulation by a catabolite control protein A (CcpA)-mediated carbon catabolite repression (CCR) mechanism, possibly with the EIIman being indirectly involved.
Conclusions: Our data shows that the ribose uptake and catabolic machinery in L. sakei is highly regulated at the transcription level. A global regulation mechanism seems to permit a fine tuning of the expression of enzymes that control efficient exploitation of available carbon sources.
Figures
References
-
- Hammes WP, Bantleon A, Min S. Lactic acid bacteria in meat fermentation. FEMS Microbiol Rev. 1990;87:165–174.
-
- Hammes WP, Hertel C. New developments in meat starter cultures. Meat Science. 1998;49:125–138. - PubMed
-
- Bredholt S, Nesbakken T, Holck A. Protective cultures inhibit growth of Listeria monocytogenes and Escherichia coli O157:H7 in cooked, sliced, vacuum- and gas-packaged meat. Int J Food Microbiol. 1999;53:43–52. - PubMed
-
- Bredholt S, Nesbakken T, Holck A. Industrial application of an antilisterial strain of Lactobacillus sakei as a protective culture and its effect on the sensory acceptability of cooked, sliced, vacuum-packaged meats. Int J Food Microbiol. 2001;66:191–196. - PubMed
-
- Katikou P, Georgantelis D, Paleologos EK, Ambrosiadis I, Kontominas MG. Relation of biogenic amines' formation with microbiological and sensory attributes in Lactobacillus-inoculated vacuum-packed rainbow trout (Oncorhynchus mykiss) fillets. J Agric Food Chem. 2006;54:4277–4283. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

