Comparative expression profiling of E. coli and S. aureus inoculated primary mammary gland cells sampled from cows with different genetic predispositions for somatic cell score
- PMID: 21702919
- PMCID: PMC3143085
- DOI: 10.1186/1297-9686-43-24
Comparative expression profiling of E. coli and S. aureus inoculated primary mammary gland cells sampled from cows with different genetic predispositions for somatic cell score
Abstract
Background: During the past ten years many quantitative trait loci (QTL) affecting mastitis incidence and mastitis related traits like somatic cell score (SCS) were identified in cattle. However, little is known about the molecular architecture of QTL affecting mastitis susceptibility and the underlying physiological mechanisms and genes causing mastitis susceptibility. Here, a genome-wide expression analysis was conducted to analyze molecular mechanisms of mastitis susceptibility that are affected by a specific QTL for SCS on Bos taurus autosome 18 (BTA18). Thereby, some first insights were sought into the genetically determined mechanisms of mammary gland epithelial cells influencing the course of infection.
Methods: Primary bovine mammary gland epithelial cells (pbMEC) were sampled from the udder parenchyma of cows selected for high and low mastitis susceptibility by applying a marker-assisted selection strategy considering QTL and molecular marker information of a confirmed QTL for SCS in the telomeric region of BTA18. The cells were cultured and subsequently inoculated with heat-inactivated mastitis pathogens Escherichia coli and Staphylococcus aureus, respectively. After 1, 6 and 24 h, the cells were harvested and analyzed using the microarray expression chip technology to identify differences in mRNA expression profiles attributed to genetic predisposition, inoculation and cell culture.
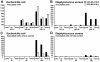
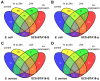
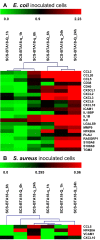
Results: Comparative analysis of co-expression profiles clearly showed a faster and stronger response after pathogen challenge in pbMEC from less susceptible animals that inherited the favorable QTL allele 'Q' than in pbMEC from more susceptible animals that inherited the unfavorable QTL allele 'q'. Furthermore, the results highlighted RELB as a functional and positional candidate gene and related non-canonical Nf-kappaB signaling as a functional mechanism affected by the QTL. However, in both groups, inoculation resulted in up-regulation of genes associated with the Ingenuity pathways 'dendritic cell maturation' and 'acute phase response signaling', whereas cell culture affected biological processes involved in 'cellular development'.
Conclusions: The results indicate that the complex expression profiling of pathogen challenged pbMEC sampled from cows inheriting alternative QTL alleles is suitable to study genetically determined molecular mechanisms of mastitis susceptibility in mammary epithelial cells in vitro and to highlight the most likely functional pathways and candidate genes underlying the QTL effect.
Figures



References
-
- Kossaibati MA, Esslemont RJ. The costs of production diseases in dairy herds in England. Vet J. 1997;154:41–51. - PubMed
-
- Seegers H, Fourichon C, Beaudeau F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet Res. 2003;34:475–491. - PubMed
-
- Tao W, Mallard B. Differentially expressed genes associated with Staphylococcus aureus mastitis of Canadian Holstein cows. Vet Immunol Immunopathol. 2007;120:201–211. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

