The HIV-1 Rev/RRE system is required for HIV-1 5' UTR cis elements to augment encapsidation of heterologous RNA into HIV-1 viral particles
- PMID: 21702950
- PMCID: PMC3131246
- DOI: 10.1186/1742-4690-8-51
The HIV-1 Rev/RRE system is required for HIV-1 5' UTR cis elements to augment encapsidation of heterologous RNA into HIV-1 viral particles
Abstract
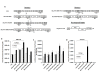
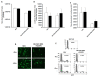
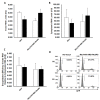
Background: The process of HIV-1 genomic RNA (gRNA) encapsidation is governed by a number of viral encoded components, most notably the Gag protein and gRNA cis elements in the canonical packaging signal (ψ). Also implicated in encapsidation are cis determinants in the R, U5, and PBS (primer binding site) from the 5' untranslated region (UTR). Although conventionally associated with nuclear export of HIV-1 RNA, there is a burgeoning role for the Rev/RRE in the encapsidation process. Pleiotropic effects exhibited by these cis and trans viral components may confound the ability to examine their independent, and combined, impact on encapsidation of RNA into HIV-1 viral particles in their innate viral context. We systematically reconstructed the HIV-1 packaging system in the context of a heterologous murine leukemia virus (MLV) vector RNA to elucidate a mechanism in which the Rev/RRE system is central to achieving efficient and specific encapsidation into HIV-1 viral particles.
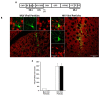
Results: We show for the first time that the Rev/RRE system can augment RNA encapsidation independent of all cis elements from the 5' UTR (R, U5, PBS, and ψ). Incorporation of all the 5' UTR cis elements did not enhance RNA encapsidation in the absence of the Rev/RRE system. In fact, we demonstrate that the Rev/RRE system is required for specific and efficient encapsidation commonly associated with the canonical packaging signal. The mechanism of Rev/RRE-mediated encapsidation is not a general phenomenon, since the combination of the Rev/RRE system and 5' UTR cis elements did not enhance encapsidation into MLV-derived viral particles. Lastly, we show that heterologous MLV RNAs conform to transduction properties commonly associated with HIV-1 viral particles, including in vivo transduction of non-dividing cells (i.e. mouse neurons); however, the cDNA forms are episomes predominantly in the 1-LTR circle form.
Conclusions: Premised on encapsidation of a heterologous RNA into HIV-1 viral particles, our findings define a functional HIV-1 packaging system as comprising the 5' UTR cis elements, Gag, and the Rev/RRE system, in which the Rev/RRE system is required to make the RNA amenable to the ensuing interaction between Gag and the canonical packaging signal for subsequent encapsidation.
Figures



Similar articles
-
The HIV-1 Rev protein enhances encapsidation of unspliced and spliced, RRE-containing lentiviral vector RNA.PLoS One. 2012;7(11):e48688. doi: 10.1371/journal.pone.0048688. Epub 2012 Nov 1. PLoS One. 2012. PMID: 23133650 Free PMC article.
-
Influence of gag and RRE Sequences on HIV-1 RNA Packaging Signal Structure and Function.J Mol Biol. 2018 Jul 6;430(14):2066-2079. doi: 10.1016/j.jmb.2018.05.029. Epub 2018 May 19. J Mol Biol. 2018. PMID: 29787767 Free PMC article.
-
Human immunodeficiency virus type 2 lentiviral vectors: packaging signal and splice donor in expression and encapsidation.J Gen Virol. 2001 Feb;82(Pt 2):425-434. doi: 10.1099/0022-1317-82-2-425. J Gen Virol. 2001. PMID: 11161282
-
On the Selective Packaging of Genomic RNA by HIV-1.Viruses. 2016 Sep 12;8(9):246. doi: 10.3390/v8090246. Viruses. 2016. PMID: 27626441 Free PMC article. Review.
-
Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles.Virology. 2014 Apr;454-455:362-70. doi: 10.1016/j.virol.2014.01.019. Epub 2014 Feb 14. Virology. 2014. PMID: 24530126 Free PMC article. Review.
Cited by
-
Multi-Faceted Post-Transcriptional Functions of HIV-1 Rev.Biology (Basel). 2012 Jul 23;1(2):165-74. doi: 10.3390/biology1020165. Biology (Basel). 2012. PMID: 24832222 Free PMC article.
-
Diverse activities of viral cis-acting RNA regulatory elements revealed using multicolor, long-term, single-cell imaging.Mol Biol Cell. 2017 Feb 1;28(3):476-487. doi: 10.1091/mbc.E16-08-0612. Epub 2016 Nov 30. Mol Biol Cell. 2017. PMID: 27903772 Free PMC article.
-
Role of Nucleocytoplasmic RNA Transport during the Life Cycle of Retroviruses.Front Microbiol. 2012 May 18;3:179. doi: 10.3389/fmicb.2012.00179. eCollection 2012. Front Microbiol. 2012. PMID: 22783232 Free PMC article.
-
Potential biomarkers in Japanese encephalitis from different hosts and geographical locations.Bioinformation. 2023 May 31;19(5):611-622. doi: 10.6026/97320630019611. eCollection 2023. Bioinformation. 2023. PMID: 37886150 Free PMC article.
-
Subcellular Localization of HIV-1 gag-pol mRNAs Regulates Sites of Virion Assembly.J Virol. 2017 Feb 28;91(6):e02315-16. doi: 10.1128/JVI.02315-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28053097 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

