In vivo versus in vitro protein abundance analysis of Shigella dysenteriae type 1 reveals changes in the expression of proteins involved in virulence, stress and energy metabolism
- PMID: 21702961
- PMCID: PMC3136414
- DOI: 10.1186/1471-2180-11-147
In vivo versus in vitro protein abundance analysis of Shigella dysenteriae type 1 reveals changes in the expression of proteins involved in virulence, stress and energy metabolism
Abstract
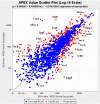
Background: Shigella dysenteriae serotype 1 (SD1) causes the most severe form of epidemic bacillary dysentery. Quantitative proteome profiling of Shigella dysenteriae serotype 1 (SD1) in vitro (derived from LB cell cultures) and in vivo (derived from gnotobiotic piglets) was performed by 2D-LC-MS/MS and APEX, a label-free computationally modified spectral counting methodology.
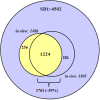
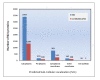
Results: Overall, 1761 proteins were quantitated at a 5% FDR (false discovery rate), including 1480 and 1505 from in vitro and in vivo samples, respectively. Identification of 350 cytoplasmic membrane and outer membrane (OM) proteins (38% of in silico predicted SD1 membrane proteome) contributed to the most extensive survey of the Shigella membrane proteome reported so far. Differential protein abundance analysis using statistical tests revealed that SD1 cells switched to an anaerobic energy metabolism under in vivo conditions, resulting in an increase in fermentative, propanoate, butanoate and nitrate metabolism. Abundance increases of transcription activators FNR and Nar supported the notion of a switch from aerobic to anaerobic respiration in the host gut environment. High in vivo abundances of proteins involved in acid resistance (GadB, AdiA) and mixed acid fermentation (PflA/PflB) indicated bacterial survival responses to acid stress, while increased abundance of oxidative stress proteins (YfiD/YfiF/SodB) implied that defense mechanisms against oxygen radicals were mobilized. Proteins involved in peptidoglycan turnover (MurB) were increased, while β-barrel OM proteins (OmpA), OM lipoproteins (NlpD), chaperones involved in OM protein folding pathways (YraP, NlpB) and lipopolysaccharide biosynthesis (Imp) were decreased, suggesting unexpected modulations of the outer membrane/peptidoglycan layers in vivo. Several virulence proteins of the Mxi-Spa type III secretion system and invasion plasmid antigens (Ipa proteins) required for invasion of colonic epithelial cells, and release of bacteria into the host cell cytosol were increased in vivo.
Conclusions: Global proteomic profiling of SD1 comparing in vivo vs. in vitro proteomes revealed differential expression of proteins geared towards survival of the pathogen in the host gut environment, including increased abundance of proteins involved in anaerobic energy respiration, acid resistance and virulence. The immunogenic OspC2, OspC3 and IpgA virulence proteins were detected solely under in vivo conditions, lending credence to their candidacy as potential vaccine targets.
Figures


References
-
- Niyogi SK. Shigellosis. J Microbiol. 2005;43(2):133–143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

