Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers
- PMID: 21702990
- PMCID: PMC3135530
- DOI: 10.1186/1472-6750-11-70
Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers
Abstract
Background: MicroRNAs are important regulators of gene expression at the post-transcriptional level and play an important role in many biological processes. Due to the important biological role it is of great interest to quantitatively determine their expression level in different biological settings.
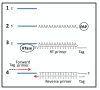
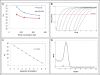
Results: We describe a PCR method for quantification of microRNAs based on a single reverse transcription reaction for all microRNAs combined with real-time PCR with two, microRNA-specific DNA primers. Primer annealing temperatures were optimized by adding a DNA tail to the primers and could be designed with a success rate of 94%. The method was able to quantify synthetic templates over eight orders of magnitude and readily discriminated between microRNAs with single nucleotide differences. Importantly, PCR with DNA primers yielded significantly higher amplification efficiencies of biological samples than a similar method based on locked nucleic acids-spiked primers, which is in agreement with the observation that locked nucleic acid interferes with efficient amplification of short templates. The higher amplification efficiency of DNA primers translates into higher sensitivity and precision in microRNA quantification.
Conclusions: MiR-specific quantitative RT-PCR with DNA primers is a highly specific, sensitive and accurate method for microRNA quantification.
Figures



References
-
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

