In vivo and in vitro function of human UDP-galactose 4'-epimerase variants
- PMID: 21703329
- PMCID: PMC3168732
- DOI: 10.1016/j.biochi.2011.06.009
In vivo and in vitro function of human UDP-galactose 4'-epimerase variants
Abstract
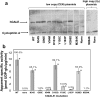
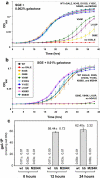
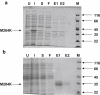
Type III galactosemia results from reduced activity of the enzyme UDP-galactose 4'-epimerase. Five disease-associated alleles (G90E, V94M, D103G, N34S and L183P) and three artificial alleles (Y105C, N268D, and M284K) were tested for their ability to alleviate galactose-induced growth arrest in a Saccharomyces cerevisiae strain which lacks endogenous UDP-galactose 4'-epimerase. For all of these alleles, except M284K, the ability to alleviate galactose sensitivity was correlated with the UDP-galactose 4'-epimerase activity detected in cell extracts. The M284K allele, however, was able to substantially alleviate galactose sensitivity, but demonstrated near-zero activity in cell extracts. Recombinant expression of the corresponding protein in Escherichia coli resulted in a protein with reduced enzymatic activity and reduced stability towards denaturants in vitro. This lack of stability may result from the introduction of an unpaired positive charge into a bundle of three α-helices near the surface of the protein. The disparities between the in vivo and in vitro data for M284K-hGALE further suggest that there are additional, stabilising factors present in the cell. Taken together, these results reinforce the need for care in the interpretation of in vitro, enzymatic diagnostic tests for type III galactosemia.
Copyright © 2011 Elsevier Masson SAS. All rights reserved.
Figures


Similar articles
-
Characterization of two mutations associated with epimerase-deficiency galactosemia, by use of a yeast expression system for human UDP-galactose-4-epimerase.Am J Hum Genet. 1997 Sep;61(3):590-8. doi: 10.1086/515517. Am J Hum Genet. 1997. PMID: 9326324 Free PMC article.
-
Identification and characterization of a mutation, in the human UDP-galactose-4-epimerase gene, associated with generalized epimerase-deficiency galactosemia.Am J Hum Genet. 1999 Feb;64(2):462-70. doi: 10.1086/302263. Am J Hum Genet. 1999. PMID: 9973283 Free PMC article.
-
Altered cofactor binding affects stability and activity of human UDP-galactose 4'-epimerase: implications for type III galactosemia.Biochim Biophys Acta. 2012 Oct;1822(10):1516-26. doi: 10.1016/j.bbadis.2012.05.007. Epub 2012 May 18. Biochim Biophys Acta. 2012. PMID: 22613355 Free PMC article.
-
The molecular basis of galactosemia - Past, present and future.Gene. 2016 Sep 10;589(2):133-41. doi: 10.1016/j.gene.2015.06.077. Epub 2015 Jul 2. Gene. 2016. PMID: 26143117 Review.
-
UDP-hexose 4-epimerases: a view on structure, mechanism and substrate specificity.Carbohydr Res. 2015 Sep 23;414:8-14. doi: 10.1016/j.carres.2015.06.006. Epub 2015 Jun 21. Carbohydr Res. 2015. PMID: 26162744 Review.
Cited by
-
Misfolding of galactose 1-phosphate uridylyltransferase can result in type I galactosemia.Biochim Biophys Acta. 2013 Aug;1832(8):1279-93. doi: 10.1016/j.bbadis.2013.04.004. Epub 2013 Apr 11. Biochim Biophys Acta. 2013. PMID: 23583749 Free PMC article.
-
GALE Promotes the Proliferation and Migration of Glioblastoma Cells and Is Regulated by miR-let-7i-5p.Cancer Manag Res. 2019 Dec 16;11:10539-10554. doi: 10.2147/CMAR.S221585. eCollection 2019. Cancer Manag Res. 2019. PMID: 31908526 Free PMC article.
-
Inherited thrombocytopenia associated with mutation of UDP-galactose-4-epimerase (GALE).Hum Mol Genet. 2019 Jan 1;28(1):133-142. doi: 10.1093/hmg/ddy334. Hum Mol Genet. 2019. PMID: 30247636 Free PMC article.
-
Galactosemia: Towards Pharmacological Chaperones.J Pers Med. 2021 Feb 7;11(2):106. doi: 10.3390/jpm11020106. J Pers Med. 2021. PMID: 33562227 Free PMC article. Review.
-
Metabolic precision labeling enables selective probing of O-linked N-acetylgalactosamine glycosylation.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25293-25301. doi: 10.1073/pnas.2007297117. Epub 2020 Sep 28. Proc Natl Acad Sci U S A. 2020. PMID: 32989128 Free PMC article.
References
-
- Leloir L.F. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch. Biochem. 1951;33:186–190. - PubMed
-
- Thoden J.B., Wohlers T.M., Fridovich-Keil J.L., Holden H.M. Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J. Biol. Chem. 2001;276:15131–15136. - PubMed
-
- Gitzelmann R., Steinmann B., Mitchell B., Haigis E. Uridine diphosphate galactose 4′-epimerase deficiency. IV. Report of eight cases in three families. Helv. Paediatr. Acta. 1977;31:441–452. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

