Compartmentation of membrane processes and nucleotide dynamics in diffusion-restricted cardiac cell microenvironment
- PMID: 21704043
- PMCID: PMC3264845
- DOI: 10.1016/j.yjmcc.2011.06.007
Compartmentation of membrane processes and nucleotide dynamics in diffusion-restricted cardiac cell microenvironment
Abstract
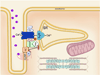
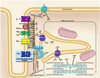
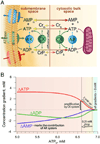
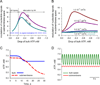
Orchestrated excitation-contraction coupling in heart muscle requires adequate spatial arrangement of systems responsible for ion movement and metabolite turnover. Co-localization of regulatory and transporting proteins into macromolecular complexes within an environment of microanatomical cell components raises intracellular diffusion barriers that hamper the mobility of metabolites and signaling molecules. Compared to substrate diffusion in the cytosol, diffusional restrictions underneath the sarcolemma are much larger and could impede ion and nucleotide movement by a factor of 10(3)-10(5). Diffusion barriers thus seclude metabolites within the submembrane space enabling rapid and vectorial effector targeting, yet hinder energy supply from the bulk cytosolic space implicating the necessity for a shunting transfer mechanism. Here, we address principles of membrane protein compartmentation, phosphotransfer enzyme-facilitated interdomain energy transfer, and nucleotide signal dynamics at the subsarcolemma-cytosol interface. This article is part of a Special Issue entitled "Local Signaling in Myocytes".
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Alberts B, Johnson A, Lewis J, Raff R, Roberts K, Walter P. Molecular Biology of the Cell. 4th edition. New York: Garland Science; 2002. The compartmentalization of cells; pp. 659–669.
-
- Rockman HA, Koch WJ, Lefkowitz RJ. Seven-transmembrane-spanning receptors and heart function. Nature. 2002;415:206–212. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

