Distinction of thioredoxin transnitrosylation and denitrosylation target proteins by the ICAT quantitative approach
- PMID: 21704743
- PMCID: PMC3253718
- DOI: 10.1016/j.jprot.2011.06.001
Distinction of thioredoxin transnitrosylation and denitrosylation target proteins by the ICAT quantitative approach
Abstract
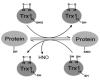
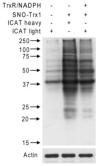
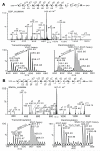
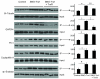
S-Nitrosylation is a reversible PTM for regulating protein function. Thioredoxin-1 (Trx1) catalyzes either transnitrosylation or denitrosylation of specific proteins, depending on the redox status of the cysteines within its conserved oxidoreductase CXXC motif. With a disulfide bond formed between the two catalytic cysteines, Trx1 is not only inactive as a denitrosylase, but it may also be nitrosylated at Cys73 and serve as a transnitrosylating agent. Identification of Trx1-mediated transnitrosylation or denitrosylation targets will contribute to a better understanding of Trx1's function. Previous experimental approaches based on the attenuation of CXXC oxidoreductase activity cannot readily distinguish Trx1 transnitrosylation targets from denitrosylation targets. In this study, we used the ICAT method in conjunction with the biotin switch technique to differentiate Trx1 transnitrosylation targets from denitrosylation target proteins from neuroblastoma cells. We demonstrate that the ICAT approach is effective for quantitative identification of putative Trx1 transnitrosylation and denitrosylation target peptides. From these analyses, we confirmed reports that peroxiredoxin 1 is a Trx1 transnitrosylation, but not a denitrosylation target, and we found several other proteins, including cyclophilin A to be modulated in this manner. Unexpectedly, we found that many nitrosylation sites are reversibly regulated by Trx1, suggesting a more prominent role for Trx1 in regulating S-nitrosylation.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Redox regulatory mechanism of transnitrosylation by thioredoxin.Mol Cell Proteomics. 2010 Oct;9(10):2262-75. doi: 10.1074/mcp.M110.000034. Epub 2010 Jul 21. Mol Cell Proteomics. 2010. PMID: 20660346 Free PMC article.
-
Functional proteomics approaches for the identification of transnitrosylase and denitrosylase targets.Methods. 2013 Aug 1;62(2):151-60. doi: 10.1016/j.ymeth.2013.02.002. Epub 2013 Feb 18. Methods. 2013. PMID: 23428400 Free PMC article.
-
Thioredoxin 1-mediated post-translational modifications: reduction, transnitrosylation, denitrosylation, and related proteomics methodologies.Antioxid Redox Signal. 2011 Nov 1;15(9):2565-604. doi: 10.1089/ars.2010.3831. Epub 2011 Jun 8. Antioxid Redox Signal. 2011. PMID: 21453190 Free PMC article. Review.
-
Small changes huge impact: the role of thioredoxin 1 in the regulation of apoptosis by S-nitrosylation.Acta Biochim Biophys Sin (Shanghai). 2013 Mar;45(3):153-61. doi: 10.1093/abbs/gms103. Epub 2012 Dec 4. Acta Biochim Biophys Sin (Shanghai). 2013. PMID: 23212077 Review.
-
Master redox regulator Trx1 upregulates SMYD1 & modulates lysine methylation.Biochim Biophys Acta. 2015 Dec;1854(12):1816-1822. doi: 10.1016/j.bbapap.2015.09.006. Epub 2015 Sep 26. Biochim Biophys Acta. 2015. PMID: 26410624 Free PMC article.
Cited by
-
Strategies for profiling native S-nitrosylation.Biopolymers. 2014 Feb;101(2):173-9. doi: 10.1002/bip.22342. Biopolymers. 2014. PMID: 23828013 Free PMC article. Review.
-
Redox Signaling Mediated by Thioredoxin and Glutathione Systems in the Central Nervous System.Antioxid Redox Signal. 2017 Nov 1;27(13):989-1010. doi: 10.1089/ars.2016.6925. Epub 2017 May 18. Antioxid Redox Signal. 2017. PMID: 28443683 Free PMC article. Review.
-
GSHSite: exploiting an iteratively statistical method to identify s-glutathionylation sites with substrate specificity.PLoS One. 2015 Apr 7;10(4):e0118752. doi: 10.1371/journal.pone.0118752. eCollection 2015. PLoS One. 2015. PMID: 25849935 Free PMC article.
-
Quantitative site-specific reactivity profiling of S-nitrosylation in mouse skeletal muscle using cysteinyl peptide enrichment coupled with mass spectrometry.Free Radic Biol Med. 2013 Apr;57:68-78. doi: 10.1016/j.freeradbiomed.2012.12.010. Epub 2012 Dec 28. Free Radic Biol Med. 2013. PMID: 23277143 Free PMC article.
-
Identification of Bax-interacting proteins in oligodendrocyte progenitors during glutamate excitotoxicity and perinatal hypoxia-ischemia.ASN Neuro. 2013 Dec 23;5(5):e00131. doi: 10.1042/AN20130027. ASN Neuro. 2013. PMID: 24195677 Free PMC article.
References
-
- Furchgott RF, Zawadzki JV. The obligatory role of endothelial cells in the relaxation of arterial smooth muscle by acetylcholine. Nature. 1980;288:373–376. - PubMed
-
- Gao Y. The multiple actions of NO. Pflugers Arch. 2010;459:829–839. - PubMed
-
- Lipton SA, Choi YB, Takahashi H, Zhang D, Li W, Godzik A, et al. Cysteine regulation of protein function—as exemplified by NMDA-receptor modulation. Trends Neurosci. 2002;25:474–480. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

