Viruses with more than 1,000 genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of Mimivirus genes
- PMID: 21705471
- PMCID: PMC3163472
- DOI: 10.1093/gbe/evr048
Viruses with more than 1,000 genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of Mimivirus genes
Abstract
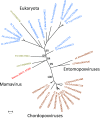
The genome sequence of the Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, is reported. With 1,191,693 nt in length and 1,023 predicted protein-coding genes, the Mamavirus has the largest genome among the known viruses. The genomes of the Mamavirus and the previously described Mimivirus are highly similar in both the protein-coding genes and the intergenic regions. However, the Mamavirus contains an extra 5'-terminal segment that encompasses primarily disrupted duplicates of genes present elsewhere in the genome. The Mamavirus also has several unique genes including a small regulatory polyA polymerase subunit that is shared with poxviruses. Detailed analysis of the protein sequences of the two Mimiviruses led to a substantial amendment of the functional annotation of the viral genomes.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

