Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4?
- PMID: 21705644
- PMCID: PMC3160039
- DOI: 10.1105/tpc.111.086264
Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4?
Abstract
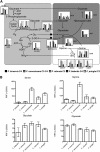
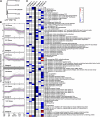
Selective pressure exerted by a massive decline in atmospheric CO(2) levels 55 to 40 million years ago promoted the evolution of a novel, highly efficient mode of photosynthetic carbon assimilation known as C(4) photosynthesis. C(4) species have concurrently evolved multiple times in a broad range of plant families, and this multiple and parallel evolution of the complex C(4) trait indicates a common underlying evolutionary mechanism that might be elucidated by comparative analyses of related C(3) and C(4) species. Here, we use mRNA-Seq analysis of five species within the genus Flaveria, ranging from C(3) to C(3)-C(4) intermediate to C(4) species, to quantify the differences in the transcriptomes of closely related plant species with varying degrees of C(4)-associated characteristics. Single gene analysis defines the C(4) cycle enzymes and transporters more precisely and provides new candidates for yet unknown functions as well as identifies C(4) associated pathways. Molecular evidence for a photorespiratory CO(2) pump prior to the establishment of the C(4) cycle-based CO(2) pump is provided. Cluster analysis defines the upper limit of C(4)-related gene expression changes in mature leaves of Flaveria as 3582 alterations.
Figures




Comment in
-
How to make a C4 plant: insight from comparative transcriptome analysis.Plant Cell. 2011 Jun;23(6):2009. doi: 10.1105/tpc.111.230612. Epub 2011 Jun 24. Plant Cell. 2011. PMID: 21705641 Free PMC article. No abstract available.
References
-
- Aoki N., Ohnishi J., Kanai R. (1992). Two different mechanisms for transport of pyruvate into mesophyll chloroplasts of C4 plants–A comparative study. Plant Cell Physiol. 33: 805–809
-
- Audic S., Claverie J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7: 986–995 - PubMed
-
- Bauwe H. (2011). Photorespiration: The bridge to C4 photosynthesis. C4 Photosynthesis and Related CO2 Concentrating Mechanisms, Raghavendra A.S., Sage R.F., (Dordrecht, The Netherlands: Springer; ), pp. 81–108
-
- Bauwe H., Keerberg O., Bassüner R., Pärnik T., Bassüner B. (1987). Reassimilation of carbon dioxide by Flaveria (Asteraceae) species representing different types of photosynthesis. Planta 172: 214–218 - PubMed
-
- Beligni M.V., Mayfield S.P. (2008). Arabidopsis thaliana mutants reveal a role for CSP41a and CSP41b, two ribosome-associated endonucleases, in chloroplast ribosomal RNA metabolism. Plant Mol. Biol. 67: 389–401 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

