Five phosphonate operon gene products as components of a multi-subunit complex of the carbon-phosphorus lyase pathway
- PMID: 21705661
- PMCID: PMC3136323
- DOI: 10.1073/pnas.1104922108
Five phosphonate operon gene products as components of a multi-subunit complex of the carbon-phosphorus lyase pathway
Abstract
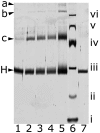
Organophosphonate utilization by Escherichia coli requires the 14 cistrons of the phnCDEFGHIJKLMNOP operon, of which the carbon-phosphorus lyase has been postulated to consist of the seven polypeptides specified by phnG to phnM. A 5,660-bp DNA fragment encompassing phnGHIJKLM is cloned, followed by expression in E. coli and purification of Phn-polypeptides. PhnG, PhnH, PhnI, PhnJ, and PhnK copurify as a protein complex by ion-exchange, size-exclusion, and affinity chromatography. The five polypeptides also comigrate in native-PAGE. Cross-linking of the purified protein complex reveals a close proximity of PhnG, PhnI, PhnJ, and PhnK, as these subunits disappear concomitant with the formation of large cross-linked protein complexes. Two molecular forms are identified, a major form of molecular mass of approximately 260 kDa, a minor form of approximately 640 kDa. The stoichiometry of the protein complex is suggested to be PhnG(4)H(2)I(2)J(2)K. Deletion of individual phn genes reveals that a strain harboring plasmid-borne phnGHIJ produces a protein complex consisting of PhnG, PhnH, PhnI, and PhnJ, whereas a strain harboring plasmid-borne phnGIJK produces a protein complex consisting of PhnG and PhnI. We conclude that phnGHIJK specify a soluble multisubunit protein complex essential for organophosphonate utilization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chen CM, Ye QZ, Zhu ZM, Wanner BL, Walsh CT. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J Biol Chem. 1990;265:4461–4471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

