Autoregulatory circuit of human rpL3 expression requires hnRNP H1, NPM and KHSRP
- PMID: 21705779
- PMCID: PMC3177206
- DOI: 10.1093/nar/gkr461
Autoregulatory circuit of human rpL3 expression requires hnRNP H1, NPM and KHSRP
Erratum in
-
Corrigendum: Autoregulatory circuit of human rpL3 expression requires hnRNP H1, NPM and KHSRP.Nucleic Acids Res. 2017 Dec 1;45(21):12600. doi: 10.1093/nar/gkx1099. Nucleic Acids Res. 2017. PMID: 29087476 Free PMC article. No abstract available.
Abstract
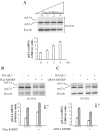
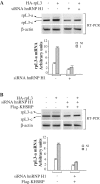
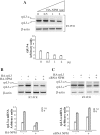
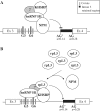
Alternative pre-mRNA splicing (AS) is a major mechanism that allows proteomic variability in eukaryotic cells. However, many AS events result in mRNAs containing a premature termination codon, which are degraded by nonsense-mediated mRNA decay (NMD) pathway. We have previously demonstrated that human rpL3 autoregulates its expression through the association of AS with NMD. In fact, overexpression of rpL3 promotes downregulation of canonical splicing and upregulation of alternative splicing that produces an NMD-targeted mRNA isoform. The result of these events is a decreased production of rpL3. We have also identified heterogeneous nuclear ribonucleoprotein (hnRNP) H1 as a splicing factor involved in the regulation of rpL3 alternative splicing and identified its regulatory cis-elements within intron 3 transcript. Here, we report that NPM and KHSRP are two newly identified proteins involved in the regulation of rpL3 gene expression via AS-NMD. We demonstrate that hnRNP H1, KHSRP and NPM can be found associated, and present also in ribonucleoproteins (RNPs) including rpL3 and intron 3 RNA in vivo, and describe protein-protein and RNA-protein interactions. Moreover, our data provide an insight on the crucial role of hnRNP H1 in the regulation of the alternative splicing of the rpL3 gene.
Figures



Comment in
-
Editor's Note to 'Autoregulatory circuit of human rpL3 expression requires hnRNP H1, NPM and KHSRP'.Nucleic Acids Res. 2022 Nov 11;50(20):11999. doi: 10.1093/nar/gkac1099. Nucleic Acids Res. 2022. PMID: 36330946 Free PMC article. No abstract available.
References
-
- Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 2001;65:289–308. - PubMed
-
- Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. - PubMed
-
- Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. - PubMed
-
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. - PubMed
-
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 2007;623:123–147. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

