Validation of a gene expression-based subclassification strategy for pediatric septic shock
- PMID: 21705885
- PMCID: PMC3196776
- DOI: 10.1097/CCM.0b013e3182257675
Validation of a gene expression-based subclassification strategy for pediatric septic shock
Abstract
Objective: Septic shock heterogeneity has important implications for clinical trial implementation and patient management. We previously addressed this heterogeneity by identifying three putative subclasses of children with septic shock based exclusively on a 100-gene expression signature. Here we attempted to prospectively validate the existence of these gene expression-based subclasses in a validation cohort.
Design: Prospective observational study involving microarray-based bioinformatics.
Setting: Multiple pediatric intensive care units in the United States.
Patients: Separate derivation (n = 98) and validation (n = 82) cohorts of children with septic shock.
Interventions: None other than standard care.
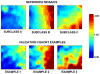
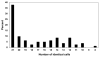
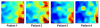
Measurements and main results: Gene expression mosaics of the 100 class-defining genes were generated for 82 individual patients in the validation cohort. Using computer-based image analysis, patients were classified into one of three subclasses ("A," "B," or "C") based on color and pattern similarity relative to reference mosaics generated from the original derivation cohort. After subclassification, the clinical database was mined for phenotyping. Subclass A patients had higher illness severity relative to subclasses B and C as measured by maximal organ failure, fewer intensive care unit-free days, and a higher Pediatric Risk of Mortality score. Patients in subclass A were characterized by repression of genes corresponding to adaptive immunity and glucocorticoid receptor signaling. Separate subclass assignments were conducted by 21 individual clinicians using visual inspection. The consensus classification of the clinicians had modest agreement with the computer algorithm.
Conclusions: We have validated the existence of subclasses of children with septic shock based on a biologically relevant, 100-gene expression signature. The subclasses have relevant clinical differences.
Conflict of interest statement
Dr. Thomas consulted for Discovery Laboratories. The remaining authors have not disclosed any potential conflicts of interest.
Figures
Comment in
-
Novel diagnostics for sepsis: a decade of promise for gene expression profiling.Crit Care Med. 2011 Nov;39(11):2579-81. doi: 10.1097/CCM.0b013e31822a5e36. Crit Care Med. 2011. PMID: 22005238 No abstract available.
References
-
- Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348(2):138–150. - PubMed
-
- Marshall JC. Sepsis: rethinking the approach to clinical research. J Leukoc Biol. 2008;83(3):471–482. - PubMed
-
- Marshall JC, Reinhart K. Biomarkers of sepsis. Crit Care Med. 2009;37(7):2290–2298. - PubMed
-
- Wong HR. Pediatric septic shock treatment: new clues from genomic profiling. Pharmacogenomics. 2007;8(10):1287–1290. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

