Compare the differences of synonymous codon usage between the two species within cardiovirus
- PMID: 21708006
- PMCID: PMC3148564
- DOI: 10.1186/1743-422X-8-325
Compare the differences of synonymous codon usage between the two species within cardiovirus
Abstract
Background: Cardioviruses are positive-strand RNA viruses in the Picornaviridae family that can cause enteric infection in rodents and also been detected at lower frequencies in other mammals such as pigs and human beings. The Cardiovirus genus consists two distinct species: Encephalomyocarditis virus (EMCV) and Theilovirus (ThV). There are a lot differences between the two species. In this study, the differences of codon usage in EMCV and ThV were compared.
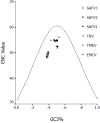
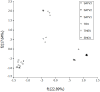
Results: The mean ENC values of EMCV and ThV are 54.86 and 51.08 respectively, higher than 40.And there are correlations between (C+G)₁₂% and (C+G)₃% for both EMCV and ThV (r = -0.736; r = 0.986, P < 0.01, repectively). For ThV the (C+G)₁₂%, (C+G)₃%, axis f'₁ and axis f'₂ had a significant correlations respectively but not for EMCV. According to the RSCU values, the EMCV species seemed to prefer U, G and C ending codon, while the ThV spice seemed to like using U and A ending codon. However, in both genus AGA for Arg, AUU for Ile, UCU for Ser, and GGA for Gly were chosen preferentially. Correspondence analysis detected one major trend in the first axis (f'₁) which accounted for 22.89% of the total variation, and another major trend in the second axis (f'₂) which accounted for 17.64% of the total variation. And the plots of the same serotype seemed at the same region at the coordinate.
Conclusion: The overall extents of codon usage bias in both EMCV and ThV are low. The mutational pressure is the main factor that determines the codon usage bias, but the (C+G) content plays a more important role in codon usage bias for ThV than for EMCV. The synonymous codon usage pattern in both EMCV and ThV genes is gene function and geography specific, but not host specific. Maybe the serotype is one factor effected the codon bias for ThV, and location has no significant effect on the variations of synonymous codon usage in these virus genes.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

