Intrasubunit and intersubunit interactions controlling assembly of active synaptic complexes during Hin-catalyzed DNA recombination
- PMID: 21708172
- PMCID: PMC3156330
- DOI: 10.1016/j.jmb.2011.06.021
Intrasubunit and intersubunit interactions controlling assembly of active synaptic complexes during Hin-catalyzed DNA recombination
Abstract
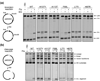
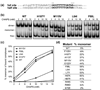
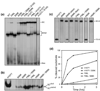
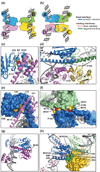
Serine recombinases, which generate double-strand breaks in DNA, must be carefully regulated to ensure that chemically active DNA complexes are assembled correctly. In the Hin-catalyzed site-specific DNA inversion reaction, two inversely oriented recombination sites on the same DNA molecule assemble into a synaptic complex that uniquely generates inversion products. The Fis-bound recombinational enhancer, together with topological constraints directed by DNA supercoiling, functions to regulate Hin synaptic complex formation and activity. We have isolated a collection of gain-of-function mutants in 22 positions within the catalytic and oligomerization domains of Hin using two genetic screens and by site-directed mutagenesis. One genetic screen measured recombination in the absence of Fis and the other assessed SOS induction as a readout of increased DNA cleavage. These mutations, together with molecular modeling, identify important sites of dynamic intrasubunit and intersubunit interactions that regulate assembly of the active tetrameric recombination complex. Of particular interest are interactions between the oligomerization helix (helix E) and the catalytic domain of the same subunit that function to hold the dimer in an inactive state in the absence of the Fis/enhancer system. Among these is a relay involving a triad of phenylalanines that are proposed to switch positions during the transition from dimers to the catalytically active tetramer. Novel Hin mutants that generate synaptic complexes that are blocked at steps prior to DNA cleavage are also described.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
The Hin dimer interface is critical for Fis-mediated activation of the catalytic steps of site-specific DNA inversion.Curr Biol. 1996 Feb 1;6(2):163-77. doi: 10.1016/s0960-9822(02)00449-9. Curr Biol. 1996. PMID: 8673463
-
Multiple interfaces between a serine recombinase and an enhancer control site-specific DNA inversion.Elife. 2013 Oct 22;2:e01211. doi: 10.7554/eLife.01211. Elife. 2013. PMID: 24151546 Free PMC article.
-
Mechanical constraints on Hin subunit rotation imposed by the Fis/enhancer system and DNA supercoiling during site-specific recombination.Mol Cell. 2009 Jun 26;34(6):746-59. doi: 10.1016/j.molcel.2009.05.020. Mol Cell. 2009. PMID: 19560425 Free PMC article.
-
The Fis protein: it's not just for DNA inversion anymore.Mol Microbiol. 1992 Nov;6(22):3257-65. doi: 10.1111/j.1365-2958.1992.tb02193.x. Mol Microbiol. 1992. PMID: 1484481 Review.
-
Site-specific DNA Inversion by Serine Recombinases.Microbiol Spectr. 2015 Feb;3(1):MDNA3-0047-2014. doi: 10.1128/microbiolspec.MDNA3-0047-2014. Microbiol Spectr. 2015. PMID: 26104558 Review.
Cited by
-
Controlling tetramer formation, subunit rotation and DNA ligation during Hin-catalyzed DNA inversion.Nucleic Acids Res. 2015 Jul 27;43(13):6459-72. doi: 10.1093/nar/gkv565. Epub 2015 Jun 8. Nucleic Acids Res. 2015. PMID: 26056171 Free PMC article.
-
The site-specific integration reaction of Listeria phage A118 integrase, a serine recombinase.Mob DNA. 2013 Jan 3;4(1):2. doi: 10.1186/1759-8753-4-2. Mob DNA. 2013. PMID: 23282060 Free PMC article.
-
Serine Resolvases.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0045-2014. doi: 10.1128/microbiolspec.MDNA3-0045-2014. Microbiol Spectr. 2015. PMID: 26104713 Free PMC article. Review.
-
Controlled rotation mechanism of DNA strand exchange by the Hin serine recombinase.Sci Rep. 2016 Apr 1;6:23697. doi: 10.1038/srep23697. Sci Rep. 2016. PMID: 27032966 Free PMC article.
-
Site-specific DNA Inversion by Serine Recombinases.Microbiol Spectr. 2015 Feb 19;3(3):1-36. doi: 10.1128/microbiolspec.MDNA3-0047-2014. Microbiol Spectr. 2015. PMID: 25844275 Free PMC article.
References
-
- Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. - PubMed
-
- Johnson RC. Bacterial Site-specific DNA inversion systems. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, D.C.: ASM Press; 2002. pp. 230–271.
-
- Zieg J, Silverman M, Hilmen M, Simon M. Recombinational switch for gene expression. Science. 1977;196:170–172. - PubMed
-
- Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. - PubMed
-
- Johnson RC, Simon MI. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985;41:781–791. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

