Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors
- PMID: 21708987
- PMCID: PMC3165464
- DOI: 10.1128/IAI.05226-11
Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors
Abstract
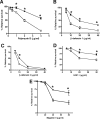
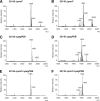
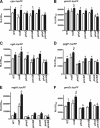
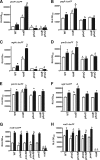
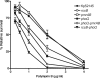
Antimicrobial peptides (APs) impose a threat to the survival of pathogens, and it is reasonable to postulate that bacteria have developed strategies to counteract them. Polymyxins are becoming the last resort to treat infections caused by multidrug-resistant Gram-negative bacteria and, similar to APs, they interact with the anionic lipopolysaccharide. Given that polymyxins and APs share the initial target, it is possible that bacterial defense mechanisms against polymyxins will be also effective against host APs. We sought to determine whether exposure to polymyxin will increase Klebsiella pneumoniae resistance to host APs. Indeed, exposure of K. pneumoniae to polymyxin induces cross-resistance not only to polymyxin itself but also to APs present in the airways. Polymyxin treatment upregulates the expression of the capsule polysaccharide operon and the loci required to modify the lipid A with aminoarabinose and palmitate with a concomitant increase in capsule and lipid A species containing such modifications. Moreover, these surface changes contribute to APs resistance and also to polymyxin-induced cross-resistance to APs. Bacterial loads of lipid A mutants in trachea and lungs of intranasally infected mice were lower than those of wild-type strain. PhoPQ, PmrAB, and the Rcs system govern polymyxin-induced transcriptional changes, and there is a cross talk between PhoPQ and the Rcs system. Our findings support the notion that Klebsiella activates a defense program against APs that is controlled by three signaling systems. Therapeutic strategies directed to prevent the activation of this program could be a new approach worth exploring to facilitate the clearance of the pathogen from the airways.
Figures


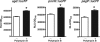

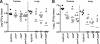
Similar articles
-
A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence.EMBO Mol Med. 2017 Apr;9(4):430-447. doi: 10.15252/emmm.201607336. EMBO Mol Med. 2017. PMID: 28202493 Free PMC article.
-
Molecular basis of Yersinia enterocolitica temperature-dependent resistance to antimicrobial peptides.J Bacteriol. 2012 Jun;194(12):3173-88. doi: 10.1128/JB.00308-12. Epub 2012 Apr 13. J Bacteriol. 2012. PMID: 22505678 Free PMC article.
-
Capsule polysaccharide is a bacterial decoy for antimicrobial peptides.Microbiology (Reading). 2008 Dec;154(Pt 12):3877-3886. doi: 10.1099/mic.0.2008/022301-0. Microbiology (Reading). 2008. PMID: 19047754
-
Mechanisms of Polymyxin Resistance.Adv Exp Med Biol. 2019;1145:55-71. doi: 10.1007/978-3-030-16373-0_5. Adv Exp Med Biol. 2019. PMID: 31364071 Review.
-
[Advances in the diagnosis and treatment strategy of polymyxin resistant Klebsiella pneumoniae].Zhonghua Jie He He Hu Xi Za Zhi. 2023 Aug 12;46(8):813-818. doi: 10.3760/cma.j.cn112147-20230418-00188. Zhonghua Jie He He Hu Xi Za Zhi. 2023. PMID: 37536993 Review. Chinese.
Cited by
-
Klebsiella pneumoniae Lipopolysaccharides Serotype O2afg Induce Poor Inflammatory Immune Responses Ex Vivo.Microorganisms. 2021 Jun 17;9(6):1317. doi: 10.3390/microorganisms9061317. Microorganisms. 2021. PMID: 34204279 Free PMC article.
-
High-throughput suppressor screen demonstrates that RcsF monitors outer membrane integrity and not Bam complex function.Proc Natl Acad Sci U S A. 2021 Aug 10;118(32):e2100369118. doi: 10.1073/pnas.2100369118. Proc Natl Acad Sci U S A. 2021. PMID: 34349021 Free PMC article.
-
Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids.Sci Rep. 2017 Jul 25;7(1):6403. doi: 10.1038/s41598-017-04793-4. Sci Rep. 2017. PMID: 28743946 Free PMC article.
-
Pharmacology of polymyxins: new insights into an 'old' class of antibiotics.Future Microbiol. 2013 Jun;8(6):711-24. doi: 10.2217/fmb.13.39. Future Microbiol. 2013. PMID: 23701329 Free PMC article. Review.
-
Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone.J Clin Microbiol. 2014 Jul;52(7):2530-5. doi: 10.1128/JCM.00088-14. Epub 2014 May 7. J Clin Microbiol. 2014. PMID: 24808234 Free PMC article.
References
-
- Agerberth B., et al. 1999. Antibacterial components in bronchoalveolar lavage fluid from healthy individuals and sarcoidosis patients. Am. J. Respir. Crit. Care Med. 160:283–290 - PubMed
-
- Aussel L., et al. 2000. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 465:87–92 - PubMed
-
- Bader M. W., et al. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219–230 - PubMed
-
- Bader M. W., et al. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

