PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment
- PMID: 21709020
- PMCID: PMC3165554
- DOI: 10.1128/MCB.05164-11
PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment
Abstract
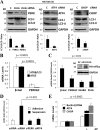
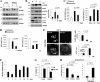
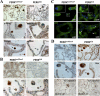
Mammary epithelial cells (MECs) detached from the extracellular matrix (ECM) produce deleterious reactive oxygen species (ROS) and induce autophagy to survive. The coordination of such opposing responses likely dictates whether epithelial cells survive ECM detachment or undergo anoikis. Here, we demonstrate that the endoplasmic reticulum kinase PERK facilitates survival of ECM-detached cells by concomitantly promoting autophagy, ATP production, and an antioxidant response. Loss-of-function studies show that ECM detachment activates a canonical PERK-eukaryotic translation initiation factor 2α (eIF2α)-ATF4-CHOP pathway that coordinately induces the autophagy regulators ATG6 and ATG8, sustains ATP levels, and reduces ROS levels to delay anoikis. Inducible activation of an Fv2E-ΔNPERK chimera by persistent activation of autophagy and reduction of ROS results in lumen-filled mammary epithelial acini. Finally, luminal P-PERK and LC3 levels are reduced in PERK-deficient mammary glands, whereas they are increased in human breast ductal carcinoma in situ (DCIS) versus normal breast tissues. We propose that the normal proautophagic and antioxidant PERK functions may be hijacked to promote the survival of ECM-detached tumor cells in DCIS lesions.
Figures





References
-
- Barone M. V., Crozat A., Tabaee A., Philipson L., Ron D. 1994. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 8:453–464 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
